













| General Information: |
|
| Name(s) found: |
MRS1 /
YIR021W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
Group I intron splicing
[IDA |
| Molecular Function: |
RNA binding
[IDA [NOT]endodeoxyribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPKNITRSV IPAIDLYCRK ANFKTLKSLS MILGSKKEWY DTKKAPLRTF LVSRCGIFEQ 60
61 LRGRLVEDGK VNLFSVFLTN DSFSFCKMTV DDKFNTSLVD WQKIPFDSTF ATDRRQNISL 120
121 LPVDTLFATE KIISILGVSP NMTNLVSIER ERSDLVDFNC KLQSNILEHL LYAKCQGVYV 180
181 TSTNEKARLL AAVCNPEFID TFWCELTPIR VSLKENPSIS VPREYQMYDP VVRATIKEVV 240
241 TKRLLRSAFD NDIDPLMCLH LDKGWKLKFP ILSSTTGLNF SLKDCLSLDT GKDASDMTEV 300
301 FLATMESSKV LRTYSNLVDI VMKDNGRLDS GVLKQFNDYV KQEKLNLQHF QAGSSKFLKG 360
361 AKI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
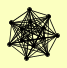
|
BI4 CBP6 MRS1 MRS2 MRS3 MRS4 MSS1 MSS116 MSS18 MTF2 NAM2 PET54 TIM10 |
| View Details | Krogan NJ, et al. (2006) |
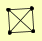
|
MRS1 RPL42B, RPL42A SEO1 SPE2 |
| View Details | Qiu et al. (2008) |
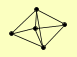
|
BI3 BI4 MRS1 MRS3 PET54 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
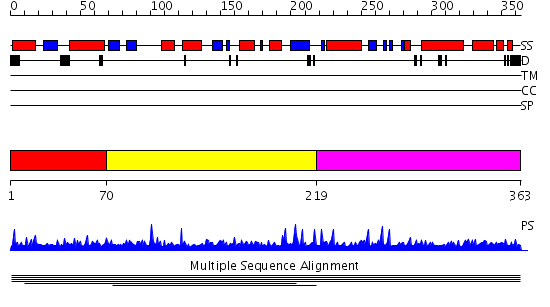
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [70..218] | 4.0 | Mitochondrial resolvase ydc2 N-terminal domain; Mitochondrial resolvase ydc2 catalytic domain | |
| 3 | View Details | [219..363] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)