













| General Information: |
|
| Name(s) found: |
BRR2 /
YER172C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2163 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U4/U6 x U5 tri-snRNP complex
[IDA U5 snRNP [IPI |
| Biological Process: |
RNA splicing
[IEA]
mRNA metabolic process [RCA] auxin biosynthetic process [IEA] mRNA processing [IEA] spliceosome conformational change to release U4 (or U4atac) and U1 (or U11) [IMP] |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEHETKDKA KKIREIYRYD EMSNKVLKVD KRFMNTSQNP QRDAEISQPK SMSGRISAKD 60
61 MGQGLCNNIN KGLKENDVAV EKTGKSASLK KIQQHNTILN SSSDFRLHYY PKDPSNVETY 120
121 EQILQWVTEV LGNDIPHDLI IGTADIFIRQ LKENEENEDG NIEERKEKIQ HELGINIDSL 180
181 KFNELVKLMK NITDYETHPD NSNKQAVAIL ADDEKSDEEE VTEMSNNANV LGGEINDNED 240
241 DDEEYDYNDV EVNSKKKNKR ALPNIENDII KLSDSKTSNI ESVPIYSIDE FFLQRKLRSE 300
301 LGYKDTSVIQ DLSEKILNDI ETLEHNPVAL EQKLVDLLKF ENISLAEFIL KNRSTIFWGI 360
361 RLAKSTENEI PNLIEKMVAK GLNDLVEQYK FRETTHSKRE LDSGDDQPQS SEAKRTKFSN 420
421 PAIPPVIDLE KIKFDESSKL MTVTKVSLPE GSFKRVKPQY DEIHIPAPSK PVIDYELKEI 480
481 TSLPDWCQEA FPSSETTSLN PIQSKVFHAA FEGDSNMLIC APTGSGKTNI ALLTVLKALS 540
541 HHYNPKTKKL NLSAFKIVYI APLKALVQEQ VREFQRRLAF LGIKVAELTG DSRLSRKQID 600
601 ETQVLVSTPE KWDITTRNSN NLAIVELVRL LIIDEIHLLH DDRGPVLESI VARTFWASKY 660
661 GQEYPRIIGL SATLPNYEDV GRFLRVPKEG LFYFDSSFRP CPLSQQFCGI KERNSLKKLK 720
721 AMNDACYEKV LESINEGNQI IVFVHSRKET SRTATWLKNK FAEENITHKL TKNDAGSKQI 780
781 LKTEAANVLD PSLRKLIESG IGTHHAGLTR SDRSLSEDLF ADGLLQVLVC TATLAWGVNL 840
841 PAHTVIIKGT DVYSPEKGSW EQLSPQDVLQ MLGRAGRPRY DTFGEGIIIT DQSNVQYYLS 900
901 VLNQQLPIES QFVSKLVDNL NAEVVAGNIK CRNDAVNWLA YTYLYVRMLA SPMLYKVPDI 960
961 SSDGQLKKFR ESLVHSALCI LKEQELVLYD AENDVIEATD LGNIASSFYI NHASMDVYNR1020
1021 ELDEHTTQID LFRIFSMSEE FKYVSVRYEE KRELKQLLEK APIPIREDID DPLAKVNVLL1080
1081 QSYFSQLKFE GFALNSDIVF IHQNAGRLLR AMFEICLKRG WGHPTRMLLN LCKSATTKMW1140
1141 PTNCPLRQFK TCPVEVIKRL EASTVPWGDY LQLETPAEVG RAIRSEKYGK QVYDLLKRFP1200
1201 KMSVTCNAQP ITRSVMRFNI EIIADWIWDM NVHGSLEPFL LMLEDTDGDS ILYYDVLFIT1260
1261 PDIVGHEFTL SFTYELKQHN QNNLPPNFFL TLISENWWHS EFEIPVSFNG FKLPKKFPPP1320
1321 TPLLENISIS TSELGNDDFS EVFEFKTFNK IQSQVFESLY NSNDSVFVGS GKGTGKTAMA1380
1381 ELALLNHWRQ NKGRAVYINP SGEKIDFLLS DWNKRFSHLA GGKIINKLGN DPSLNLKLLA1440
1441 KSHVLLATPV QFELLSRRWR QRKNIQSLEL MIYDDAHEIS QGVYGAVYET LISRMIFIAT1500
1501 QLEKKIRFVC LSNCLANARD FGEWAGMTKS NIYNFSPSER IEPLEINIQS FKDVEHISFN1560
1561 FSMLQMAFEA SAAAAGNRNS SSVFLPSRKD CMEVASAFMK FSKAIEWDML NVEEEQIVPY1620
1621 IEKLTDGHLR APLKHGVGIL YKGMASNDER IVKRLYEYGA VSVLLISKDC SAFACKTDEV1680
1681 IILGTNLYDG AEHKYMPYTI NELLEMVGLA SGNDSMAGKV LILTSHNMKA YYKKFLIEPL1740
1741 PTESYLQYII HDTLNNEIAN SIIQSKQDCV DWFTYSYFYR RIHVNPSYYG VRDTSPHGIS1800
1801 VFLSNLVETC LNDLVESSFI EIDDTEAEVT AEVNGGDDEA TEIISTLSNG LIASHYGVSF1860
1861 FTIQSFVSSL SNTSTLKNML YVLSTAVEFE SVPLRKGDRA LLVKLSKRLP LRFPEHTSSG1920
1921 SVSFKVFLLL QAYFSRLELP VDFQNDLKDI LEKVVPLINV VVDILSANGY LNATTAMDLA1980
1981 QMLIQGVWDV DNPLRQIPHF NNKILEKCKE INVETVYDIM ALEDEERDEI LTLTDSQLAQ2040
2041 VAAFVNNYPN VELTYSLNNS DSLISGVKQK ITIQLTRDVE PENLQVTSEK YPFDKLESWW2100
2101 LVLGEVSKKE LYAIKKVTLN KETQQYELEF DTPTSGKHNL TIWCVCDSYL DADKELSFEI2160
2161 NVK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
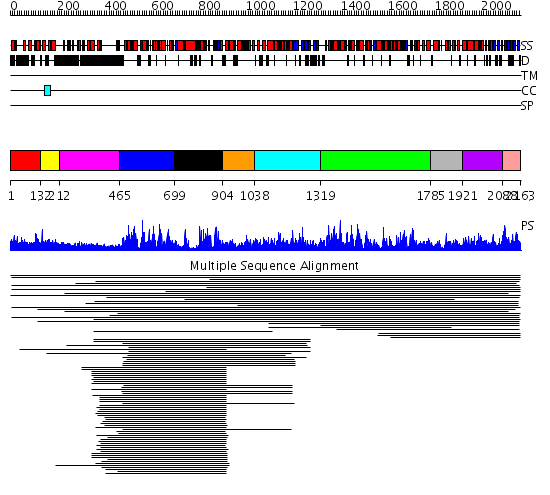
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 1.019994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [132..211] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [212..464] | 4.418997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [465..698] | 86.69897 | Putative DEAD box RNA helicase | |
| 5 | View Details | [699..903] | 86.69897 | Putative DEAD box RNA helicase | |
| 6 | View Details | [904..1037] | 5.69897 | Nucleotide excision repair enzyme UvrB | |
| 7 | View Details | [1038..1318] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1319..1784] | 72.257996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1785..1920] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1921..2087] | 3.154902 | Ribosomal protein L32e | |
| 11 | View Details | [2088..2163] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)