













| General Information: |
|
| Name(s) found: |
MRD1 /
YPR112C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 887 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [IDA |
| Biological Process: |
endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
[IMP]
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] small-subunit processome assembly [IMP] endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] rRNA processing [IEA] |
| Molecular Function: |
snoRNA binding
[IDA rRNA primary transcript binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRIIVKGLP VYLTDDNLRE HFTKRLRQKH SHQAVNGSGP DLITDVKILR DRNGESRRFG 60
61 FIGYRNEEDA FDAVEYFNGS FINTSKIEVS MAKSFADPRV PQPMKEKRRE ALKRFREKEE 120
121 KLLQEENRKK KKVDENKHSN IDDEIRKNKQ LQEFMETMKP SSQVTSWEKV GIDKSIEDEK 180
181 LKREEEDSSV QGNSLLAHAL ALKEENNKDE APNLVIENES DDEYSALNRN RDEDQEDAGE 240
241 EEKMISISNL KDTDIGLVND DANSDEKENE KRRNLAQDEK VSDLDWFKQR RVRIKESEAE 300
301 TREKSSSYAT EQNESLDTKK EEQPERAVPQ KTDEELAIEK INQTGRLFLR NILYTSKEED 360
361 FRKLFSPFGE LEEVHVALDT RTGQSKGFAY VLFKDSKNAV NAYVELDKQI FQGRLLHILP 420
421 GEEKKSHRLD EFDLKNMPLK KQKELKRKAA ASRQTFSWNS LYMNQDAVLG SVAAKLGLEK 480
481 SQLIDAENSS SAVKQALAEA HVIGDVRKYF ESKGVDLTKF SQLKSTNQRD DKVILVKNFP 540
541 FGTTREELGE MFLPYGKLER LLMPPAGTIA IVQFRDTTSA RAAFTKLSYK RFKDGIIYLE 600
601 RGPKDCFTKP AEADDLINNT SAKEEENPVE VKPSSNDLME ANKDVTEGSS NAHDEDVIDG 660
661 PTVSIFIKNL NFSTTNQNLT DRFKVFTGFV VAQVKTKPDP KHQGKTLSMG FGFVEFRTKE 720
721 QANAVIAAMD GTVIDGHKIQ LKLSHRQASQ SGNTKTKSNK KSGKIIVKNL PFEATRKDVF 780
781 ELFNSFGQLK SVRVPKKFDK SARGFAFVEF LLPKEAENAM DQLHGVHLLG RRLVMQYAEE 840
841 DAVDAEEEIA RMTKKVRKQV ATNEMAALRN GGGRKKLDVD DEENEGF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BMS1 DIP2 MRD1 UTP22 |
| View Details | Krogan NJ, et al. (2006) |
|
GDT1 MRD1 NOP12 RRP17 |
| View Details | Gavin AC, et al. (2006) |
|
BMS1 CDC36 CMS1 DIP2 MRD1 NAN1 NOP58 PNO1 PWP2 ROK1 RPS10B RRP9 UTP10 UTP13 UTP15 UTP18 UTP21 UTP22 UTP4 UTP6 UTP7 UTP8 UTP9 |
| View Details | Gavin AC, et al. (2002) |

|
ADE4 ASC1 BFR2 BMS1 BRX1 BUD21 CBF5 CKA1 CKA2 CKB1 CKB2 CMS1 DBP8 DIA4 DIM1 DIP2 DIS3 ECM16 ECM29 EMG1 ENP1 ENP2 ERO1 ESF1 ESF2 FAP7 GCN1 HCA4 HHF1, HHF2 HSL1 HTA2 IMP3 IMP4 KRE33 KRR1 LCP5 LTV1 MPP10 MRD1 MVD1 NAN1 NOB1 NOC2 NOC4 NOP1 NOP14 NOP56 NOP58 NOP6 PNO1 PRE6 PRE9 PRP43 PWP2 RCL1 RML2 ROK1 RPP2B RRP12 RRP7 RRP9 RTG2 SAM1 SCL1 SEC21 SLX9 SOF1 TIF1, TIF2 TSR1 UTP10 UTP11 UTP13 UTP15 UTP18 UTP20 UTP21 UTP22 UTP30 UTP4 UTP6 UTP7 UTP8 UTP9 YOR059C |
| View Details | Qiu et al. (2008) |
|
BRX1 DIP2 ENP1 HAS1 MRD1 NAN1 NSR1 PWP2 RRP3 RRP5 RRP9 SBP1 SOF1 UTP11 UTP15 UTP4 UTP5 UTP6 UTP7 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
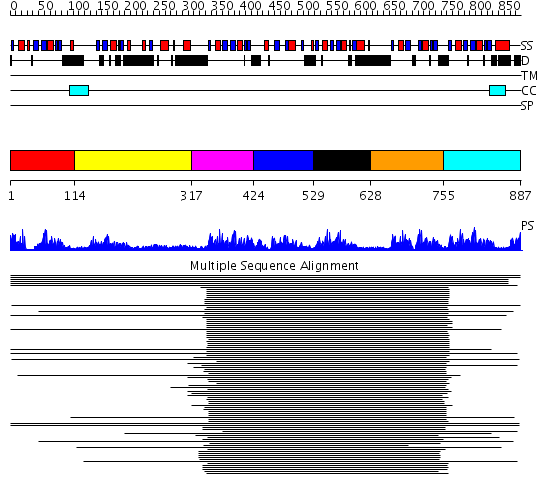
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | 7.0 | Nucleolin | |
| 2 | View Details | [114..316] | 1.132995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [317..423] | 121.232149 | Poly(A)-binding protein | |
| 4 | View Details | [424..528] | 121.232149 | Poly(A)-binding protein | |
| 5 | View Details | [529..627] | 41.980454 | Nucleolin | |
| 6 | View Details | [628..754] | 97.40824 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 7 | View Details | [755..887] | 97.40824 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)