













| General Information: |
|
| Name(s) found: |
POL1 /
YNL102W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[IDA alpha DNA polymerase:primase complex [IDA mitochondrion [IDA |
| Biological Process: |
DNA replication initiation
[IC lagging strand elongation [IC DNA replication [IMP DNA synthesis involved in DNA repair [IMP premeiotic DNA synthesis [IMP |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] DNA binding [IEA] DNA-directed DNA polymerase activity [IDA][IEA] nucleotidyltransferase activity [IEA] transferase activity [IEA] nucleoside binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKSEKLEK LRKLQAARNG TSIDDYEGDE SDGDRIYDEI DEKEYRARKR QELLHDDFVV 60
61 DDDGVGYVDR GVEEDWREVD NSSSDEDTGN LASKDSKRKK NIKREKDHQI TDMLRTQHSK 120
121 STLLAHAKKS QKKSIPIDNF DDILGEFESG EVEKPNILLP SKLRENLNSS PTSEFKSSIK 180
181 RVNGNDESSH DAGISKKVKI DPDSSTDKYL EIESSPLKLQ SRKLRYANDV QDLLDDVENS 240
241 PVVATKRQNV LQDTLLANPP SAQSLADEED DEDSDEDIIL KRRTMRSVTT TRRVNIDSRS 300
301 NPSTSPFVTA PGTPIGIKGL TPSKSLQSNT DVATLAVNVK KEDVVDPETD TFQMFWLDYC 360
361 EVNNTLILFG KVKLKDDNCV SAMVQINGLC RELFFLPREG KTPTDIHEEI IPLLMDKYGL 420
421 DNIRAKPQKM KYSFELPDIP SESDYLKVLL PYQTPKSSRD TIPSDLSSDT FYHVFGGNSN 480
481 IFESFVIQNR IMGPCWLDIK GADFNSIRNA SHCAVEVSVD KPQNITPTTT KTMPNLRCLS 540
541 LSIQTLMNPK ENKQEIVSIT LSAYRNISLD SPIPENIKPD DLCTLVRPPQ STSFPLGLAA 600
601 LAKQKLPGRV RLFNNEKAML SCFCAMLKVE DPDVIIGHRL QNVYLDVLAH RMHDLNIPTF 660
661 SSIGRRLRRT WPEKFGRGNS NMNHFFISDI CSGRLICDIA NEMGQSLTPK CQSWDLSEMY 720
721 QVTCEKEHKP LDIDYQNPQY QNDVNSMTMA LQENITNCMI SAEVSYRIQL LTLTKQLTNL 780
781 AGNAWAQTLG GTRAGRNEYI LLHEFSRNGF IVPDKEGNRS RAQKQRQNEE NADAPVNSKK 840
841 AKYQGGLVFE PEKGLHKNYV LVMDFNSLYP SIIQEFNICF TTVDRNKEDI DELPSVPPSE 900
901 VDQGVLPRLL ANLVDRRREV KKVMKTETDP HKRVQCDIRQ QALKLTANSM YGCLGYVNSR 960
961 FYAKPLAMLV TNKGREILMN TRQLAESMNL LVVYGDTDSV MIDTGCDNYA DAIKIGLGFK1020
1021 RLVNERYRLL EIDIDNVFKK LLLHAKKKYA ALTVNLDKNG NGTTVLEVKG LDMKRREFCP1080
1081 LSRDVSIHVL NTILSDKDPE EALQEVYDYL EDIRIKVETN NIRIDKYKIN MKLSKDPKAY1140
1141 PGGKNMPAVQ VALRMRKAGR VVKAGSVITF VITKQDEIDN AADTPALSVA ERAHALNEVM1200
1201 IKSNNLIPDP QYYLEKQIFA PVERLLERID SFNVVRLSEA LGLDSKKYFR REGGNNNGED1260
1261 INNLQPLETT ITDVERFKDT VTLELSCPSC DKRFPFGGIV SSNYYRVSYN GLQCKHCEQL1320
1321 FTPLQLTSQI EHSIRAHISL YYAGWLQCDD STCGIVTRQV SVFGKRCLND GCTGVMRYKY1380
1381 SDKQLYNQLL YFDSLFDCEK NKKQELKPIY LPDDLDYPKE QLTESSIKAL TEQNRELMET1440
1441 GRSVVQKYLN DCGRRYVDMT SIFDFMLN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
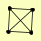
|
POL1 POL12 PRI1 PRI2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
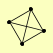
|
POL1 POL12 PRI1 PRI2 |
| View Details | Krogan NJ, et al. (2006) |
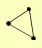
|
POL1 POL12 PRI1 |
| View Details | Gavin AC, et al. (2006) |
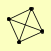
|
POL1 POL12 PRI1 PRI2 |
| View Details | Gavin AC, et al. (2002) |
|
AHA1 ATP11 CAF130 CAF40 CCR4 CCT6 CDC36 CDC39 COP1 FAS2 GCN1 MIS1 MOT2 NOT3 NOT5 PDC1 POL1 POL12 POP2 PRI1 PRI2 RFC1 RFC3 RFC4 RFC5 RVB2 SAM1 SEC27 TFC7 TFP1 USO1 YRA1 |
| View Details | Qiu et al. (2008) |
|
DPB2 DPB3 DPB4 MCM3 MCM4 MCM6 POL1 POL12 POL2 POL3 POL30 POL31 POL32 PRI1 PRI2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
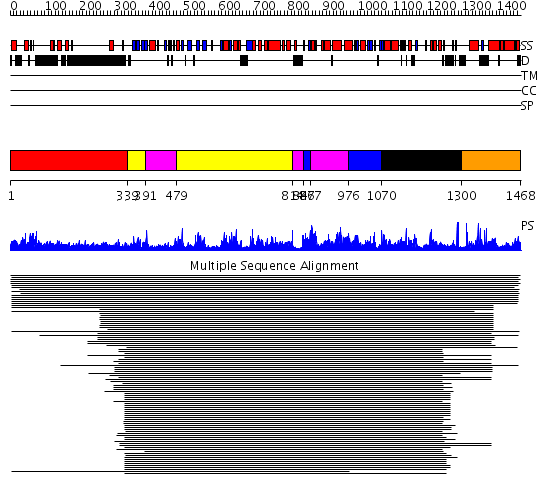
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..338] | 4.071997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [339..390] [479..813] |
1450.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 3 | View Details | [391..478] [814..846] [867..975] |
1450.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 4 | View Details | [847..866] [976..1069] |
1450.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 5 | View Details | [1070..1299] | 1450.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 6 | View Details | [1300..1468] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)