













| General Information: |
|
| Name(s) found: |
EBP2 /
YKL172W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 427 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
nuclear division
[IMP ribosome biogenesis [RCA rRNA processing [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKGFKLKEL LSHQKEIEKA EKLENDLKKK KSQELKKEEP TIVTASNLKK LEKKEKKADV 60
61 KKEVAADTEE YQSQALSKKE KRKLKKELKK MQEQDATEAQ KHMSGDEDES GDDREEEEEE 120
121 EEEEEGRLDL EKLAKSDSES EDDSESENDS EEDEDVVAKE ESEEKEEQEE EQDVPLSDVE 180
181 FDSDADVVPH HKLTVNNTKA MKHALERVQL PWKKHSFQEH QSVTSETNTD EHIKDIYDDT 240
241 ERELAFYKQS LDAVLVARDE LKRLKVPFKR PLDYFAEMVK SDEHMDKIKG KLIEEASDKK 300
301 AREEARRQRQ LKKFGKQVQN ATLQKRQLEK RETLEKIKSL KNKRKHNEID HSEFNVGVEE 360
361 EVEGKRFDRG RPNGKRAAKN AKYGQGGMKR FKRKNDATSS ADVSGFSSRK MKGKTNRPGK 420
421 SRRARRF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
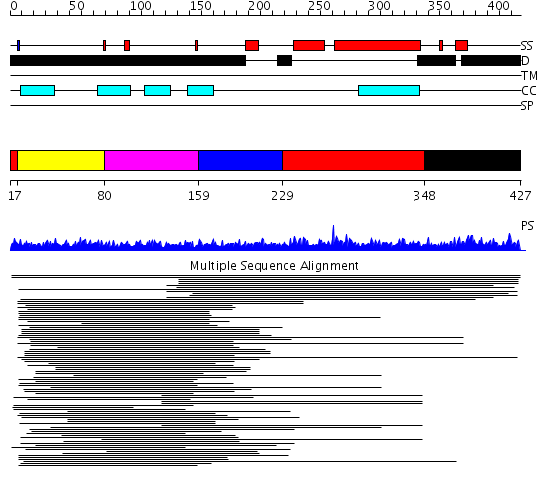
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..6] [229..347] |
10.86 | No description for 1l8mA was found. | |
| 2 | View Details | [7..79] | 10.86 | No description for 1l8mA was found. | |
| 3 | View Details | [80..158] | 10.86 | No description for 1l8mA was found. | |
| 4 | View Details | [159..228] | 10.86 | No description for 1l8mA was found. | |
| 5 | View Details | [348..427] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)