













| General Information: |
|
| Name(s) found: |
LAA1 /
YJL207C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2014 amino acids |
Gene Ontology: |
|
| Cellular Component: |
AP-1 adaptor complex
[IPI ribosome [IDA clathrin-coated vesicle [IDA |
| Biological Process: |
protein localization
[IMP retrograde transport, endosome to Golgi [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANRSLKKVI ETSSNNGHDL LTWITTNLEK LICLKEVNDN EIQEVKEIHT QLDEFVRYIS 60
61 VLENTDDLEL HSVFISLSQL YTISIWRLKD EYPGVVFDSA AFLTNVLCEE DVSIDDGDTD 120
121 PNQKKKKKKS STKKKKYIYS PAKDIACTIL VQLFENFGSS ISSLIPLLFN AIFKNLKKIM 180
181 EKSKYYHATF MTTLLQLFNA ILRNSNNDDK ILDPATYAKF SKLSKTVFDS ISTDEKDFSV 240
241 TFVSVLIECW TAHFKQTNFI REHSHDIIET IYSRFTEGEI GVYGFANDET RIFTAKSLAE 300
301 ILFDYYFSKN ILTLQEVWSI YVKIFLNCDT RDVESGCFES IIHLINLNLL ADNTFLSNSK 360
361 YLDIVLSLSG VFSSYEVNNR SMNTLSRYLR YFQHMHEVIL PHLNDSAKTQ MLYYILGCSD 420
421 TYQSSSKSDS ASNFKYSIDA KPETQWLTLL QLDFTYVLIS DLGSTFTTEE NTVKEIRDKL 480
481 VDLATCEIFT IRVHTVEILK VFLNNCPEYL SETIENSLRA LSTDFKSTGK FIFHKNHGHA 540
541 FIIANLIKGA ESDYISYELI MRITVFSTSF IKNNTTSTSS NLYFKGLLCW ILLIGLMNYK 600
601 DEQYLKLQIP QLFLFWKVLL THTYTYHDED ELYKNLEIRN HALTCLLTYL SNTTIDKEMA 660
661 KQVSYLLTKC SNFNHSIDLK SKNIDNALLH NENRILQVYL KLEKYINSDF NSSLLILIVK 720
721 NFSDPNLYTE SSSSVLGSLK DIGNRKVSNK DDMESNIVLE SSINTLLRQN NGFAFGLSSK 780
781 ITGDRIVNLS MSSAYKYDES ISGSWPSKDY NWYNIFEVEV SKPISPILSL DSLILLYGSG 840
841 SYSQIDRYAP QVTTSLIDSS MELFSSVFPF LNSKIQYSIM ETLNLSMFSK MTTPLRSVAV 900
901 AANVCSALHN ALRIMQENNL ELDYSVGQLI IESIKKIQFF NDIFLTKIKA DCVGLLTAAI 960
961 ARTLGDEERQ KFLTEQSRIF IKNVADMDEP YLRMFHVLSL ATIFKYNSQY ANFEEYFDVI1020
1021 FALMRDPHPV VHSWSLKAMH ILLEKHLVID LKTAALLLSS MEELLVQDKY GIYGRSTLRC1080
1081 NYNRDFNSHV AIGEISRTLT ETVGPNFLEL NTKVLDSFRN ITLSMLISNN ILNSITSIKM1140
1141 FENIATFKMK NILNYEIFIL ASKSIIKSSI VTGIGSSYFD TTFTGSNELI SRTSSLKGAF1200
1201 ENFDLLTLLY KLQMEEFFMK EMENLSWRYL ALFPNSGSVK NYFTEWILHT FKRDNHWFDK1260
1261 LYSIFNMSLG RLFQSYNRDV SALLEVNGLK KSSEKEIKGE EEESIANVNQ LTDTDAGGLD1320
1321 SENLQWKSRQ IILNLILMLC LESEKYENLL LALSNKIADL IKISFRGSTV RNEGMKLTGL1380
1381 HILNFVLKNY STMRDPQVPG SSILEQQEAQ ITSALMPAFS KGSSPTVMSF AITVAAEVLA1440
1441 SNIMPPDKLG RISQLLIDLL GNFKDPNSGI RIGEAIIVTP KAKRKIELAV LDAWAEVVQR1500
1501 SITSSNDALF SFTRKYWSIL VPLWIISLRE YMMIKYNDND STVQVKNDSK ENSLIEPRST1560
1561 KIELYEPVWL NFVEALGCTL DSDVQVILAS LNDEELEYFL FILFSQCLEA IVKNIDDHSV1620
1621 KMQVLPALHN VLKSNLCIKS IFEDDIITEV VEIMDRLIST GDSKEEFLLV DIISDLIIGY1680
1681 SKCNATPETF LQDIDKLYEL LRLLMTIISE RLPFIKYNVL TSEEDDNEIK ISPTDISLLK1740
1741 KTFIAFESNI SNFDNMFKVD LYSCLLFIIG KIYECSHREV IIPIILPLFK ALVKALTESE1800
1801 DEKNIVLLEI FYGSIKDVIY HKLDSKNKVA TILILLSNGY SKLSFQELNQ CANILSEALN1860
1861 NPATQPIALQ GFKRIISNIF KYPLLQYFMK LVIKRFFQDI QTNDSLSQAS IKTKLIIQFS1920
1921 EEVIKQDHQK ASLSIALCLS FFAAYHSAYT EKIDNEVASG IVALAKLDKN SFKEAISSTI1980
1981 SPQQKAIIGS VMEAYVKSQS LGSVEEAFQL KSFD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ADE2 BSC2 CWH41 DBF4 DIG1 ECM29 HAL5 LAA1 MRC1 OPI9 PEX32 PRP5 PTH2 RGA2 RMA1 RPN3 SIR1 STE12 THR1 TLG2 VPS45 YDL199C YEL057C YPL035C |
| View Details | Ho Y, et al. (2002) |
|
BCY1 CAR2 CDC33 CKA1 CKA2 CKB1 CYS4 ESF1 ESF2 GND1 GTO1 HTB1 IMD3 LAA1 LHP1 LYS1 MSS116 NOP12 PMA1 RPF2 SAH1 SNO2 SSN8 THI22 THO2 TIF1, TIF2 TPK2 TPK3 VPS21 YBR028C YCK1 YCK2 YCK3 YCR087W YDR102C YGR111W YPT53 YRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
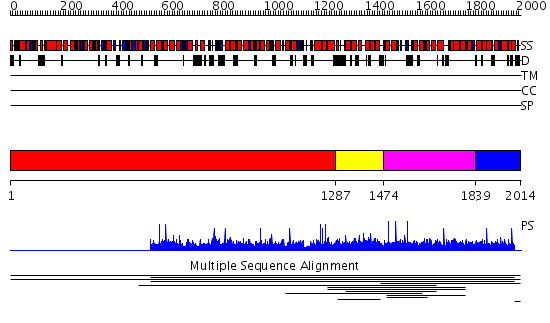
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1286] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [1287..1473] | 1.007958 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1474..1838] | 1.009949 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1839..2014] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [551..780] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [781..842] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [843..1554] | 1.19 | Adaptin beta subunit N-terminal fragment | |
| 8 | View Details | [1555..1684] | 2.05697 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1685..1802] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1803..1881] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1882..2014] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)