













| General Information: |
|
| Name(s) found: |
NAM8 /
YHR086W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[IPI U1 snRNP [IDA |
| Biological Process: |
mRNA splice site selection
[IMP nuclear mRNA splicing, via spliceosome [IPI |
| Molecular Function: |
RNA binding
[IPI mRNA binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYKQTTYYP SRGNLVRNDS SPYTNTISSE TNNSSTSVLS LQGASNVSLG TTGNQLYMGD 60
61 LDPTWDKNTV RQIWASLGEA NINVRMMWNN TLNNGSRSSM GPKNNQGYCF VDFPSSTHAA 120
121 NALLKNGMLI PNFPNKKLKL NWATSSYSNS NNSLNNVKSG NNCSIFVGDL APNVTESQLF 180
181 ELFINRYAST SHAKIVHDQV TGMSKGYGFV KFTNSDEQQL ALSEMQGVFL NGRAIKVGPT 240
241 SGQQQHVSGN NDYNRSSSSL NNENVDSRFL SKGQSFLSNG NNNMGFKRNH MSQFIYPVQQ 300
301 QPSLNHFTDP NNTTVFIGGL SSLVTEDELR AYFQPFGTIV YVKIPVGKCC GFVQYVDRLS 360
361 AEAAIAGMQG FPIANSRVRL SWGRSAKQTA LLQQAMLSNS LQVQQQQPGL QQPNYGYIPS 420
421 STCEAPVLPD NNVSSTMLPG CQILNYSNPY ANANGLGSNN FSFYSNNNAT NTQATSLLAD 480
481 TSSMDLSGTG GQQVIMQGSE AVVNSTNAML NRLEQGSNGF MFA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
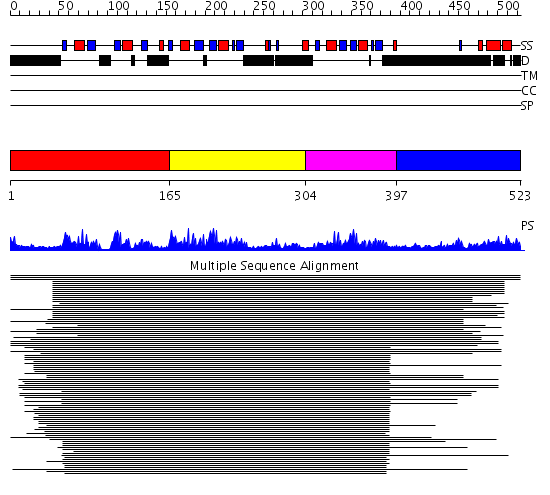
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 126.67837 | Poly(A)-binding protein | |
| 2 | View Details | [165..303] | 14.522879 | Polypyrimidine tract-binding protein | |
| 3 | View Details | [304..396] | 14.522879 | Polypyrimidine tract-binding protein | |
| 4 | View Details | [397..523] | 1.382997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)