













| General Information: |
|
| Name(s) found: |
SNU71 /
YGR013W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 620 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U1 snRNP
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IPI |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRDIVFVSPQ LYLSSQEGWK SDSAKSGFIP ILKNDLQRFQ DSLKHIVDAR NSLSETLLNS 60
61 NDDGSIHNSD QNTGLNKDKE ASIADNNSAN KCATSSSRYQ ELKQFLPISL DQQIHTVSLQ 120
121 GVSSSFSRGQ IESLLDHCLN LALTETQSNS ALKVEAWSSF SSFLDTQDIF IRFSKVDEDE 180
181 AFVNTLNYCK ALFAFIRKLH EDFKIELHLD LNTKEYVEDR TGTIPSVKPE KASEFYSVFK 240
241 NIEDQTDERN SKKEQLDDSS TQYKVDTNTL SDLPSDALDQ LCKDIIEFRT KVVSIEKEKK 300
301 MKSTYEESRR QRHQMQKVFD QIRKNHSGAK GSANTEEEDT NMEDEDEEDD TEDDLALEKR 360
361 KEERDLEESN RRYEDMLHQL HSNTEPKIKS IRADIMSAEN YEEHLEKNRS LYLKELLHLA 420
421 NDVHYDHHRS FKEQEERRDE EDRAKNGNAK ELAPIQLSDG KAISAGKAAA ITLPEGTVKS 480
481 ENYNADKNVS ESSEHVKIKF DFKKAIDHSV ESSSEDEGYR ESELPPTKPS ERSAAEDRLP 540
541 FTADELNIRL TNLKESRYVD ELVREFLGVY EDELVEYILE NIRVNQSKQA LLNELRETFD 600
601 EDGETIADRL WSRKEFRLGT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
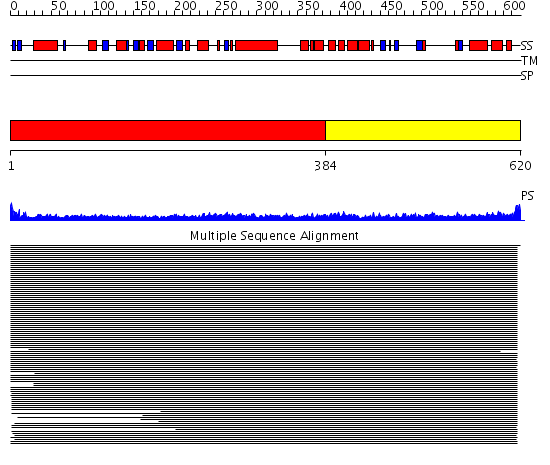
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..383] | 20.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [384..620] | 5.195997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [521..620] | 3.15 | Solution structure of the PWI motif from SRm160 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)