













| General Information: |
|
| Name(s) found: |
VPS41 /
YDR080W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 992 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA fungal-type vacuole [IDA vacuolar membrane [IDA HOPS complex [IPI |
| Biological Process: |
vacuolar protein processing
[IMP vacuole organization [IMP protein transport [IMP vacuole fusion, non-autophagic [IDA |
| Molecular Function: |
Rab guanyl-nucleotide exchange factor activity
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTDNHQNDS VLDQQSGERT IDESNSISDE NNVDNKREDV NVTSPTKSVS CISQAENGVA 60
61 SRTDESTITG SATDAETGDD DDDDDDDDDE DEDDEDEPPL LKYTRISQLP KNFFQRDSIS 120
121 SCLFGDTFFA FGTHSGILHL TTCAFEPIKT IKCHRSSILC INTDGKYFAT GSIDGTVIIG 180
181 SMDDPQNITQ YDFKRPINSV ALHSNFQASR MFVSGGMAGD VVLSQRNWLG NRIDIVLNKK 240
241 KKKKTRKDDL SSDMKGPIMG IYTMGDLILW MDDDGITFCD VPTRSQLLNI PFPSRIFNVQ 300
301 DVRPDLFRPH VHFLESDRVV IGWGSNIWLF KVSFTKDSNS IKSGDSNSQS NNMSHFNPTT 360
361 NIGSLLSSAA SSFRGTPDKK VELECHFTVS MLITGLASFK DDQLLCLGFD IDIEEEATID 420
421 EDMKEGKNFS KRPENLLAKG NAPELKIVDL FNGDEIYNDE VIMKNYEKLS INDYHLGKHI 480
481 DKTTPEYYLI SSNDAIRVQE LSLKDHFDWF MERKQYYKAW KIGKYVIGSE ERFSIGLKFL 540
541 NSLVTKKDWG TLVDHLNIIF EETLNSLDSN SYDVTQNVLK EWADIIEILI TSGNIVEIAP 600
601 LIPKKPALRK SVYDDVLHYF LANDMINKFH EYITKWDLKL FSVEDFEEEL ETRIEAASEP 660
661 TASSKEEGSN ITYRTELVHL YLKENKYTKA IPHLLKAKDL RALTIIKIQN LLPQYLDQIV 720
721 DIILLPYKGE ISHISKLSIF EIQTIFNKPI DLLFENRHTI SVARIYEIFE HDCPKSFKKI 780
781 LFCYLIKFLD TDDSFMISPY ENQLIELYSE YDRQSLLPFL QKHNNYNVES AIEVCSSKLG 840
841 LYNELIYLWG KIGETKKALS LIIDELKNPQ LAIDFVKNWG DSELWEFMIN YSLDKPNFTK 900
901 AILTCSDETS EIYLKVIRGM SDDLQIDNLQ DIIKHIVQEN SLSLEVRDNI LVIINDETKK 960
961 FANEFLKIRS QGKLFQVDES DIEINDDLNG VL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
PEP3 PEP5 VPS16 VPS33 VPS41 VPS8 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
VAM6 VPS41 |
| View Details | Gavin AC, et al. (2006) |
|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Gavin AC, et al. (2002) |
|
ATP8 LUC7 PEP3 PEP5 SNU71 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Ho Y, et al. (2002) |

|
RCK2 VPS41 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
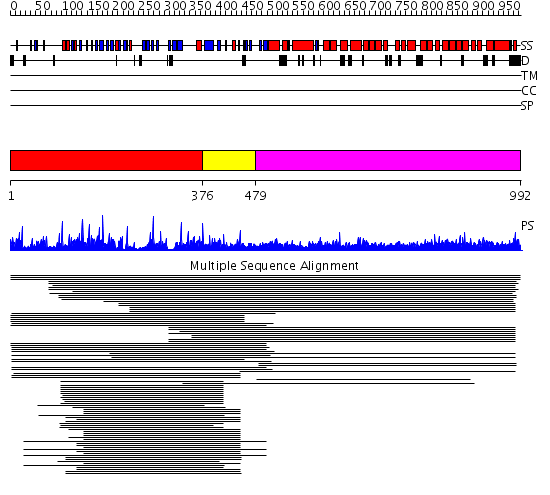
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..375] | 57.0 | Tup1, C-terminal domain | |
| 2 | View Details | [376..478] | 68.30103 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 3 | View Details | [479..992] | 12.83 | Clathrin heavy chain proximal leg segment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)