













| General Information: |
|
| Name(s) found: |
PEP5 /
YMR231W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1029 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA HOPS complex [IPI |
| Biological Process: |
vesicle docking during exocytosis
[IMP late endosome to vacuole transport [IGI vacuole fusion, non-autophagic [IDA Golgi to endosome transport [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSSWRQFQ LFENIPIRDP NFGGDSLLYS DPTLCAATIV DPQTLIIAVN SNIIKVVKLN 60
61 QSQVIHEFQS FPHDFQITFL KVINGEFLVA LAESIGKPSL IRVYKLEKLP NREQLYHSQV 120
121 ELKNGNNTYP ISVVSISNDL SCIVVGFING KIILIRGDIS RDRGSQQRII YEDPSKEPIT 180
181 ALFLNNDATA CFAATTSRIL LFNTTGRNRG RPSLVLNSKN GLDLNCGSFN PATNEFICCL 240
241 SNFIEFFSSS GKKHQFAFDL SLRKRIFCVD KDHILIVTEE TGVPTTSISV NELSPTIINR 300
301 IFIIDAKNKI ISLNFVVSSA IIDIFSTSQS GKNITYLLTS EGVMHRITPK SLENQINIII 360
361 QKELYPFALQ LAKQHSLSPL DVQEIHKKYG DYLFKKGLRK EATDQYIQCL DVVETSEIIS 420
421 KFGVKEVPDP ESMRNLADYL WSLIKNSISQ RDHVTLLLIV LIKLKDVEGI DTFIQHFDRK 480
481 GIWNEGVVMD DMDDVTFFYS DNDFFDLDLI LELMKESDFK RLSYRLAKKY SKDSLIIVDI 540
541 LLNLLHNPVK AIKYIKSLPI DETLRCLVTY SKKLLEESPN ETNALLIEVF TGKFKPSTFE 600
601 VDLDRRDTTG DFSENIRTVF YSYKTFFNYM NSNGTSDAMS ESSEASHEHE EPTYHPPKPS 660
661 IVFSSFVTKP FEFVVFLEAC LACYQQYEGF DEDRQVILTT LYDLYLNLAQ NDVPERIDDW 720
721 RSRATGVLRE SNKLVYSAAS NNTSKRVDNS IMLLISHMDQ SSASAKDKTK IDIASFANDN 780
781 PEMDLLSTFR AMTLNEEPST CLKFLEKYGT EEPKLLQVAL SYFVSNKLIF KEMGGNEVLK 840
841 EKVLRPIIEG ERMPLLDIIK ALSRTNVAHF GLIQDIIIDH VKTEDTEIKR NEKLIESYDK 900
901 ELKEKNKKLK NTINSDQPLH VPLKNQTCFM CRLTLDIPVV FFKCGHIYHQ HCLNEEEDTL 960
961 ESERKLFKCP KCLVDLETSN KLFEAQHEVV EKNDLLNFAL NSEEGSRDRF KVITEFLGRG1020
1021 AISYSDITI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
PEP3 PEP5 VPS16 VPS33 |
| View Details | Gavin AC, et al. (2006) |
|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Gavin AC, et al. (2002) |
|
ATP8 LUC7 PEP3 PEP5 SNU71 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Qiu et al. (2008) |
|
PEP3 PEP5 VPS16 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
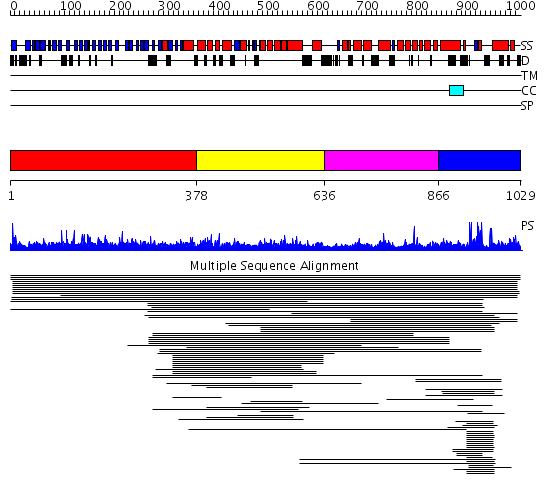
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..377] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [378..635] | 21.136677 | Region in Clathrin and VPS No confident structure predictions are available. | |
| 3 | View Details | [636..865] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [866..1029] | 2.69897 | bard1 RING domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)