













| General Information: |
|
| Name(s) found: |
VAM6 /
YDL077C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1049 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole
[IDA vacuolar membrane [IDA HOPS complex [IPI |
| Biological Process: |
vacuole organization
[IMP telomere maintenance [IMP vacuole fusion, non-autophagic [IDA |
| Molecular Function: |
Rab guanyl-nucleotide exchange factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRAQKLHSL KSSDITAILP TEQSQKLVLA KKNGDVEVYS RDGNTLKLFQ VYPDLLQNAK 60
61 NDPLPPVIEN FYFANELSTI FAQCKETLIL LSTTNLHEYD RIIDRRGINH CWLFERSHKN 120
121 KEEKNTYLIY STINTAKMRV LIWEGRTYKN MMEASLSYRK ETIRSIYPGE TGITLATDLG 180
181 IYHWPYNKPS LIRIEKTVKN KFPKDMISAL TELKEQAEKV IEKKPKKNSH FDAQSFSSMD 240
241 RMSRKSSMSS LWYRTIRNER GNKIRYTFEL DGNDATPMII DGATKKIFKV ELMHNNEEPF 300
301 LIATDHATFS ESNSEFDHMQ YLSSNLLMLY NSSTIKFVDY ENGFTFLQQK IPEGIKWVKN 360
361 LSGTYFLVWT SNDEVQLFSY HVDDGSEDDD QESICGDIND PDFYQLWRKV LFYKFFIDSP 420
421 HSKELCVSDN PEESLDICAM KLRDLTVMWC LRIFDKFQNY MVQLERSRNS RMIRSKCEEM 480
481 IIKSIFDLFI KFWAPPQLVI LKVFPSAISS LVLEITGQEH HCLLKEAEEV KETYDIPPHL 540
541 LNRWCLPYLT DTRRHLQNLL SKENDDESRI TWCYRDREIK QSFDFFLISN HDDVDLNTML 600
601 TLIDTVLFKC YLYYNPPMVG PFIRVENHCD SHVIVTELKI RHMFKDLIDF YYKRGNHEEA 660
661 LKFLTDLVDE LENDNTDQKQ RQKIDHGVKI LVIYYLKKLS NPQLDVIFTY TDWLLNRHND 720
721 SIKEILSSIF FYDSQACSSR DHLKVYGYIK KFDKLLAIQY LEFAISTFRL EGNKLHTVLI 780
781 KLYLENLDIP STRIKLKSLL ETTSVYEPRT ILKLLNDAIE SGSDQLPTNQ LNFVKYLKIF 840
841 PLSKLENHKE AVHILLDEID DYKAATSYCN DVYQSDSTKG EELLLYLYSK LVSIYDSNRN 900
901 SKLILNFLQD HGSKLNSAEI YKNLPQDISL YDIGRVVSQL LKKHTSKMDE TRLEKALLQV 960
961 ELVATTYKLN ERMSSYGVLS DSHKCPICKK VISNFGTDSI SWFTREGRNI ITHYNCGKVL1020
1021 QERFNAKNEK SSRIKQKTLG EVINELNNK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
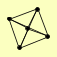
|
PEP3 PEP5 VAM6 VPS16 VPS33 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
VAM6 VPS41 |
| View Details | Krogan NJ, et al. (2006) |
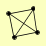
|
RLM1 TPA1 VAM6 YOR051C |
| View Details | Gavin AC, et al. (2006) |
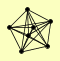
|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Gavin AC, et al. (2002) |
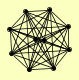
|
ATP8 LUC7 PEP3 PEP5 SNU71 VAM6 VPS16 VPS33 VPS41 VPS8 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | sample: hx1 | Hao Xu, et al. (2010) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
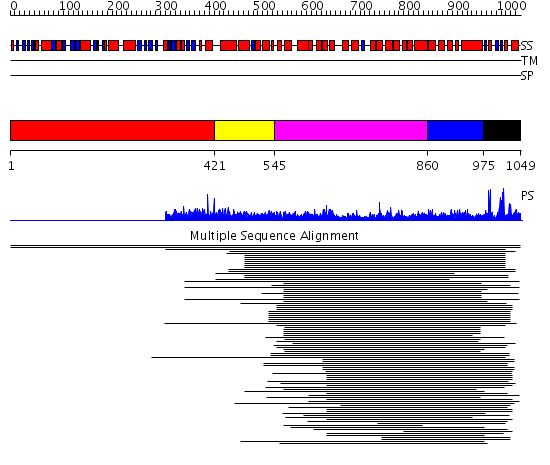
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..420] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [421..544] | 1.028988 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [545..859] | 41.045757 | Clathrin heavy chain proximal leg segment | |
| 4 | View Details | [860..974] | 1.08399 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [975..1049] | 19.090997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)