













| General Information: |
|
| Name(s) found: |
PRP28 /
YDR243C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 588 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U5 snRNP
[IPI spliceosomal complex [TAS |
| Biological Process: |
RNA splicing
[IEA]
mRNA processing [IEA] cis assembly of pre-catalytic spliceosome [TAS] |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARPIDVSQL IAGINKKKGL DENTSGKISK PRFLNKQERS KQERLKENEE SLTPTQSDSA 60
61 KVEIKKVNSR DDSFFNETND KKRNPSKQNG SKFHFSWNES EDTLSGYDPI VSTRAIDLLW 120
121 KGKTPKNAAE SSYMGKHWTE KSLHEMNERD WRILKEDYAI VTKGGTVENP LRNWEELNII 180
181 PRDLLRVIIQ ELRFPSPTPI QRITIPNVCN MKQYRDFLGV ASTGSGKTLA FVIPILIKMS 240
241 RSPPRPPSLK IIDGPKALIL APTRELVQQI QKETQKVTKI WSKESNYDCK VISIVGGHSL 300
301 EEISFSLSEG CDILVATPGR LIDSLENHLL VMKQVETLVL DEADKMIDLG FEDQVTNILT 360
361 KVDINADSAV NRQTLMFTAT MTPVIEKIAA GYMQKPVYAT IGVETGSEPL IQQVVEYADN 420
421 DEDKFKKLKP IVAKYDPPII IFINYKQTAD WLAEKFQKET NMKVTILHGS KSQEQREHSL 480
481 QLFRTNKVQI MIATNVAARG LDIPNVSLVV NFQISKKMDD YIHRIGRTGR AANEGTAVSF 540
541 VSAAEDESLI RELYKYVRKH DPLNSNIFSE AVKNKYNVGK QLSNEIIY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
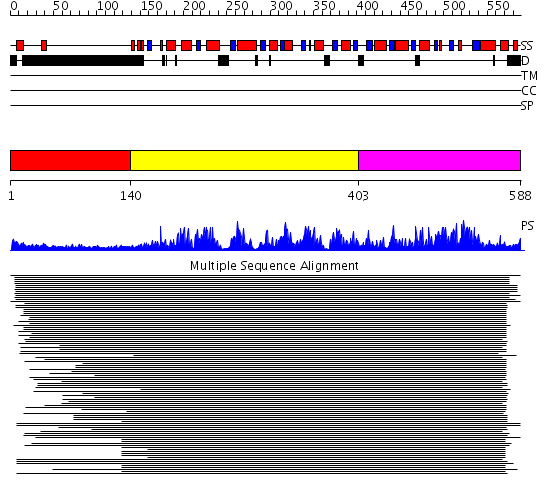
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..402] | 503.0 | Putative DEAD box RNA helicase | |
| 3 | View Details | [403..588] | 503.0 | Putative DEAD box RNA helicase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)