













| General Information: |
|
| Name(s) found: |
PAA1 /
YDR071C
[SGD]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 191 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
chromatin organization
[IMP |
| Molecular Function: |
diamine N-acetyltransferase activity
[IDA aralkylamine N-acetyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSSTLPL HMYIRPLIIE DLKQILNLES QGFPPNERAS EEIISFRLIN CPELCSGLFI 60
61 REIEGKEVKK ETLIGHIMGT KIPHEYITIE SMGKLQVESS NHIGIHSVVI KPEYQKKNLA 120
121 TLLLTDYIQK LSNQEIGNKI VLIAHEPLIP FYERVGFKII AENTNVAKDK NFAEQKWIDM 180
181 ERELIKEEYD N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
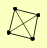
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
PAA1 PTC2 PTC3 PTC4 |
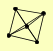
| View Details | Krogan NJ, et al. (2006) |
|
PAA1 PTC2 PTC3 PTC4 VPS34 |
| View Details | Gavin AC, et al. (2006) |

|
PAA1 PTC3 |
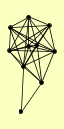
| View Details | Gavin AC, et al. (2002) |
|
ENP1 HRR25 PAA1 PFK1 PTC3 PTC4 RAD50 TSR1 VPS15 YDR186C |
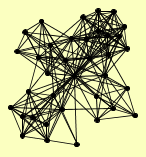
| View Details | Ho Y, et al. (2002) |
|
ADH4 ARC1 ASF1 COP1 ECM29 ENP1 GID8 GIN4 HEM15 HRP1 HTA2 IPP1 KAP104 KTR3 MDH1 MER1 NAB2 NDE1 NUM1 PAA1 PTC3 PTC4 RAD53 REP1 RPT4 SEF1 SKT5 SMC3 SPT16 SYF1 TCB1 TDA10 TPS1 UTP22 VHS1 YNL035C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
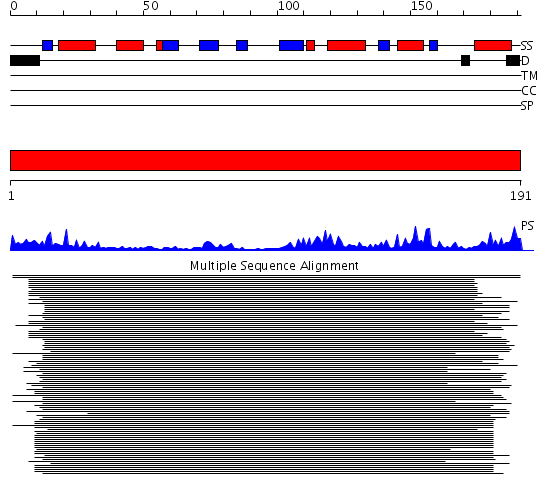
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 107.9691 | Serotonin N-acetyltranferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)