













| General Information: |
|
| Name(s) found: |
MDM1 /
YML104C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1127 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS intermediate filament [TAS |
| Biological Process: |
mitochondrion inheritance
[IDA nuclear migration [TAS mitochondrion organization [TAS |
| Molecular Function: |
structural constituent of cytoskeleton
[TAS phosphatidylinositol-3-phosphate binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKFPQFRLI LVLFYLISMI QWSVITFSLG FFLNVCIFAY FVFFKSLPDL PKPQPRFVDI 60
61 VPESSNTVDV DKELKSVEGL IQDGNAQIGK ELESIVNLII KDFVQPWFTK IDKNSDAEFL 120
121 KVIKWRLLQT LLVVKDKLMK NDSASLIVLK LLPIFNKHFS TFCDAREAVL SDLTLERHKS 180
181 ANIDLQIAVE FNKNYKIHKS LSLKPNALQK EIEKSIRKTV IGLLPHLFDN DELDSLLVFT 240
241 LMTEVLTTCI ISPLIFKFTD PDSWNLRIVS LSQNYFEEKH KVHKIRRMLS KELQDHRKVM 300
301 NDVANKDVGE PSSEKLELNA EYTGKQFEHY LNQLDSLLDL SDIKYVAYSL ALKIYQLKEN 360
361 EHLTKENLKY KKRLLLSLNL IESKLSFPGS EIDTASKKLA REANYPDLNM DNGIVLKEMA 420
421 SFLTSITLKD IVDDSEFLPF FESFLGSVPE TQGSTFLEYS QTIESFKNPL EDATSEDIIS 480
481 GYSGISTMQL QEISSKFFHN NNLQNMKLLD EGLVKNIILF RNSFQINNDE DTFILARKSV 540
541 LLLQTEAIKY LDDRFLPLFK KTPSFLKMLS TSHIISTDIY AHFLSRIGGV NNPEQNKIIK 600
601 DNVKTDFMNP VRIFANPGIT DALDNIVNGS GSKPHKSRIS SNPRYSQLFG SENDNIFKDK 660
661 LFDDENDNTS EISVVEDQLD HPRNMEKVSV SSGNSGLNPS QFYGSNNFRD NIASLTISID 720
721 QIEKELELLR HLILKADLTN NQMQLKILKK SQRTLLKELE MKELLKQQYM VQENGNSLFR 780
781 KTKIYIRSYF SENSSNGLKE ITYYIINIHH FNNGQVSSWD MARRYNEFFE LNTYLKKNFR 840
841 DLMRQLQDLF PSKVKMSLKY HVTKTLLYEE RKQKLEKYLR ELLSISEICE DNIFRRFLTD 900
901 PTPFKLNKEY MHDDILEEPL HEPIGSSNST SNSSSVVDLQ SSEDGGELNF YEDERHFFTD 960
961 SGYPFYSQNK SFVKQICDLF ISLFALNKAN AGWLRGRAII TVLQQLLGST IEKYIKVSIQ1020
1021 KLRSEDQVFE AIVTFKNMLW GDNGLFERKR NETAEATRSE GERLRTEQLA LTSLQRLFAD1080
1081 TCGRVVGLRD SHEAAGRVHA MLQNPYLNAS LLLEALDAIL LDIICND |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
ARP1 JNM1 MDM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
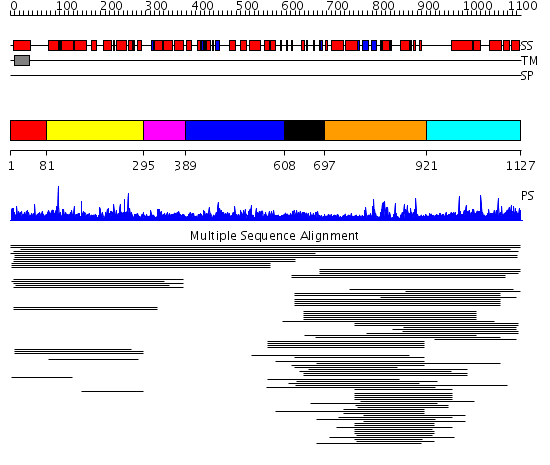
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..294] | 97.568636 | PXA domain No confident structure predictions are available. | |
| 3 | View Details | [295..388] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [389..607] | 12.29 | Regulator of G-protein signalling 4, RGS4 | |
| 5 | View Details | [608..696] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [697..920] | 22.154902 | p40phox NADPH oxidase | |
| 7 | View Details | [921..1127] | 1.075952 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|