













| General Information: |
|
| Name(s) found: |
YBL104C
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1038 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLIKKVTHW SYDNLIDYLS VNPTRDEVTH YKVDPENESD ESIIKLHTVK DFGSITCLDY 60
61 SESEIGMIGV GEKNGYLRIF NISGQNSSSP ASHAPVGLNA NNETSMTNAS GGKAAQAENI 120
121 VGSVSNLKDT QGYPVSETNY DIRVRAKKQR CINSLGINTN GLIAMGLDRN KHDSSLQIWD 180
181 MNYHDDSHET INPMFSYCTN ESIVSLKFLN DTSVLAASTK FLKEIDVRSP NPIYQHPTRL 240
241 TYDIKLNPFN DWQFSTYGDD GTLAIWDRRK LSDQASLGDL NVASPLLTFE KLVGSGAASR 300
301 KYMNSCFRWS CVRNNEFATL HRGDTIKRWR LGYYCDSNRD IAADDDNEMN IENLFVSSVH 360
361 DTNTMYDRVA TFDYIPRSNN GTSLICMRQS GTIYRMPISE VCSKAILNNR NSLLLSNFEN 420
421 TEIDEIRVNN EHEKSNLENV KTILKNLSFE DLDVSEDYFP SGHDEPNNEI EYSELSEEEN 480
481 EGSNDVLDSK RGFELFWKPE KLLEKDISVI MRTRASLGYG LDPMNTVEMI DSSKNLQNNA 540
541 YIRNTWRWIA IAKASVDDGT MVSGDLDLGY EGVIGIWNGI NGISNQDRYR QETILSDKQL 600
601 NKEMEKIIKL RRKNRDRNSP IANAAGSPKY VQRRLCLIIS GWDLSRSDYE DKYNIIMKNG 660
661 HYEKAAAWAV FFGDIPKAVE ILGSAKKERL RLIATAIAGY LAYKDLPGNN AWRQQCRKMS 720
721 SELDDPYLRV IFAFIADNDW WDILYEPAIS LRERLGVALR FLNDTDLTTF LDRTSSTVIE 780
781 NGELEGLILT GITPNGIDLL QSYVNKTSDV QSAALISIFG SPRYFRDQRV DEWIQTYRDM 840
841 LKSWELFSMR ARFDVLRSKL SRTKTGVLTA DIKPRQIYIQ CQNCKQNINT PRTSSPSSAV 900
901 STSAGNYKNG EAYRRNNADY KKFNTGSSEA QAADEKPRHK YCCPHCGSSF PRCAICLMPL 960
961 GTSNLPFVIN GTQSRDPMQT EDSQDGANRE LVSRKLKLNE WFSFCLSCNH GMHAGHAEEW1020
1021 FDRHNVCPTP GCTCQCNK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2006) |

|
CIN1 YBL104C |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
|
APM1 BMS1 BUD9 CDC53 CIC1 COF1 CTF4 DAK2 DBP10 DED81 DIA2 ENP1 FPR3 ILS1 KRE33 KRE6 KRI1 LST4 MCM2 MCM3 MCM5 NIP7 NMD3 NOP12 OMS1 RPN1 RRP12 SEH1 SKP1 SLT2 SPB1 SSF1 SSF2 SSZ1 THI22 TIF6 UFD4 URB1 UTP10 VMA6 YAK1 YBL104C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
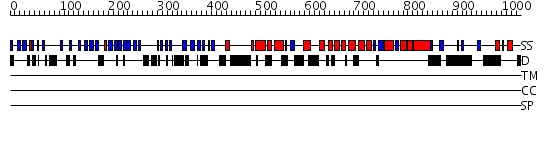
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..477] | 37.39794 | Actin interacting protein 1 | |
| 2 | View Details | [478..621] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [622..738] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [739..807] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [808..1038] | 3.72 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)