













| General Information: |
|
| Name(s) found: |
DNM1 /
YLL001W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial outer membrane [IDA |
| Biological Process: |
mitochondrion inheritance
[IGI protein homooligomerization [IDA mitochondrion organization [IMP mitochondrial fission [IGI |
| Molecular Function: |
identical protein binding
[IDA protein homodimerization activity [IDA GTPase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLEDLIPT VNKLQDVMYD SGIDTLDLPI LAVVGSQSSG KSSILETLVG RDFLPRGTGI 60
61 VTRRPLVLQL NNISPNSPLI EEDDNSVNPH DEVTKISGFE AGTKPLEYRG KERNHADEWG 120
121 EFLHIPGKRF YDFDDIKREI ENETARIAGK DKGISKIPIN LKVFSPHVLN LTLVDLPGIT 180
181 KVPIGEQPPD IEKQIKNLIL DYIATPNCLI LAVSPANVDL VNSESLKLAR EVDPQGKRTI 240
241 GVITKLDLMD SGTNALDILS GKMYPLKLGF VGVVNRSQQD IQLNKTVEES LDKEEDYFRK 300
301 HPVYRTISTK CGTRYLAKLL NQTLLSHIRD KLPDIKTKLN TLISQTEQEL ARYGGVGATT 360
361 NESRASLVLQ LMNKFSTNFI SSIDGTSSDI NTKELCGGAR IYYIYNNVFG NSLKSIDPTS 420
421 NLSVLDVRTA IRNSTGPRPT LFVPELAFDL LVKPQIKLLL EPSQRCVELV YEELMKICHK 480
481 CGSAELARYP KLKSMLIEVI SELLRERLQP TRSYVESLID IHRAYINTNH PNFLSATEAM 540
541 DDIMKTRRKR NQELLKSKLS QQENGQTNGI NGTSSISSNI DQDSAKNSDY DDDGIDAESK 600
601 QTKDKFLNYF FGKDKKGQPV FDASDKKRSI AGDGNIEDFR NLQISDFSLG DIDDLENAEP 660
661 PLTEREELEC ELIKRLIVSY FDIIREMIED QVPKAVMCLL VNYCKDSVQN RLVTKLYKET 720
721 LFEELLVEDQ TLAQDRELCV KSLGVYKKAA TLISNIL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Ho Y, et al. (2002) |
|
ABP1 ACO1 AIM41 ATP4 BUD14 CAR2 COR1 CRZ1 CYS4 DCP1 DCP2 DCS1 DNM1 EDC3 EDE1 EGD2 ENP1 GAS1 GCN3 GPH1 HHF1, HHF2 HRR25 HSP104 HXT7 HYP2 IPP1 LOC1 LSP1 LTV1 MDH1 MDS3 MGE1 NSA1 NUG1 OYE2 PEX19 PIL1 PIN4 PTC4 PUF3 RIO2 RPC19 RPM2 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SIT4 TMA19 TSR1 UBP15 VMA4 VPS1 YAK1 YBR225W YER138C YPL247C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Lockshon D, et al. (2006) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
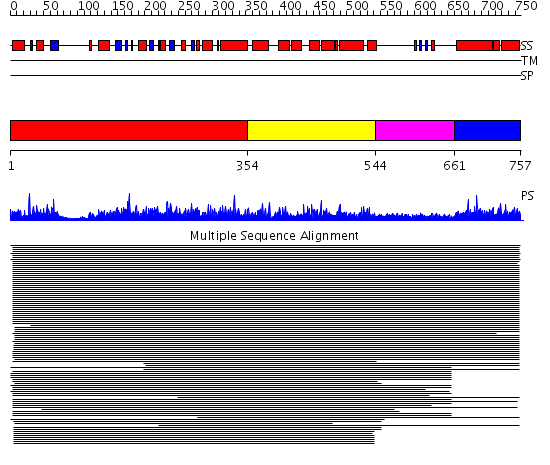
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..353] | 1175.0 | Dynamin G domain | |
| 2 | View Details | [354..543] | 50.318759 | Dynamin central region No confident structure predictions are available. | |
| 3 | View Details | [544..660] | 15.39794 | Dynamin | |
| 4 | View Details | [661..757] | 42.552842 | Dynamin GTPase effector domain Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)