













| General Information: |
|
| Name(s) found: |
VAS1 /
YGR094W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1104 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IMP mitochondrion [IDA |
| Biological Process: |
valyl-tRNA aminoacylation
[IDA |
| Molecular Function: |
valine-tRNA ligase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKWLNTLSK TFTFRLLNCH YRRSLPLCQN FSLKKSLTHN QVRFFKMSDL DNLPPVDPKT 60
61 GEVIINPLKE DGSPKTPKEI EKEKKKAEKL LKFAAKQAKK NAAATTGASQ KKPKKKKEVE 120
121 PIPEFIDKTV PGEKKILVSL DDPALKAYNP ANVESSWYDW WIKTGVFEPE FTADGKVKPE 180
181 GVFCIPAPPP NVTGALHIGH ALTIAIQDSL IRYNRMKGKT VLFLPGFDHA GIATQSVVEK 240
241 QIWAKDRKTR HDYGREAFVG KVWEWKEEYH SRIKNQIQKL GASYDWSREA FTLSPELTKS 300
301 VEEAFVRLHD EGVIYRASRL VNWSVKLNTA ISNLEVENKD VKSRTLLSVP GYDEKVEFGV 360
361 LTSFAYPVIG SDEKLIIATT RPETIFGDTA VAVHPDDDRY KHLHGKFIQH PFLPRKIPII 420
421 TDKEAVDMEF GTGAVKITPA HDQNDYNTGK RHNLEFINIL TDDGLLNEEC GPEWQGMKRF 480
481 DARKKVIEQL KEKNLYVGQE DNEMTIPTCS RSGDIIEPLL KPQWWVSQSE MAKDAIKVVR 540
541 DGQITITPKS SEAEYFHWLG NIQDWCISRQ LWWGHRCPVY FINIEGEEHD RIDGDYWVAG 600
601 RSMEEAEKKA AAKYPNSKFT LEQDEDVLDT WFSSGLWPFS TLGWPEKTKD METFYPFSML 660
661 ETGWDILFFW VTRMILLGLK LTGSVPFKEV FCHSLVRDAQ GRKMSKSLGN VIDPLDVITG 720
721 IKLDDLHAKL LQGNLDPREV EKAKIGQKES YPNGIPQCGT DAMRFALCAY TTGGRDINLD 780
781 ILRVEGYRKF CNKIYQATKF ALMRLGDDYQ PPATEGLSGN ESLVEKWILH KLTETSKIVN 840
841 EALDKRDFLT STSSIYEFWY LICDVYIENS KYLIQEGSAI EKKSAKDTLY ILLDNALKLI 900
901 HPFMPFISEE MWQRLPKRST EKAASIVKAS YPVYVSEYDD VKSANAYDLV LNITKEARSL 960
961 LSEYNILKNG KVFVESNHEE YFKTAEDQKD SIVSLIKAID EVTVVRDASE IPEGCVLQSV1020
1021 NPEVNVHLLV KGHVDIDAEI AKVQKKLEKA KKSKNGIEQT INSKDYETKA NTQAKEANKS1080
1081 KLDNTVAEIE GLEATIENLK RLKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
HEM13 PSK1 VAS1 |
| View Details | Gavin AC, et al. (2002) |

|
ACC1 ADA2 ADH1 CCT8 CLU1 CMR1 CPA2 DNA2 FDH1 FUN19 GCN20 GCN5 GFA1 GUS1 HDA1 HDA2 HDA3 HEM13 HFI1 HSC82 ILV1 KAP114 KAP123 KAP95 LCD1 LYS12 MEC1 MED4 MES1 MGM101 MLH1 MNN4 MOT1 MPH1 MRPL3 MSH2 MSH3 MSH6 MYO1 MYO4 NGG1 NOP1 OLA1 PFK2 PGK1 PMS1 PRO2 PSA1 PSK1 PTC1 PUF6 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RPA135 RPB2 RPN10 RPN11 RPN12 RPN8 RPN9 RPO21 RPT3 RPT5 RPT6 RSC8 RVB2 SAM1 SEC27 SGF29 SGF73 SGS1 SHS1 SIN3 SIN4 SMC3 SPT15 SPT20 SPT7 SPT8 TAF1 TAF10 TAF11 TAF12 TAF13 TAF14 TAF2 TAF3 TAF4 TAF5 TAF6 TAF7 TAF8 TAF9 TBF1 TCB1 TEF4 TIF1, TIF2 TOP3 TSA1 TUB3 UBP8 URA7 VAS1 VPS1 YHR122W YOL098C |
| View Details | Ho Y, et al. (2002) |
|
ADK1 ATP3 ATP5 ATP7 CDC14 CMD1 CMK2 CMP2 COF1 CPR1 DPM1 EDE1 FUR1 GLC7 HCH1 HEF3 HMS1 HYP2 ILS1 IPP1 MCR1 MYO2 MYO4 MYO5 PGM2 PST2 SHE3 SNF4 SOD1 SPE3 SSZ1 TPS1 TSA2 UBA1 VAS1 VPS13 YNK1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
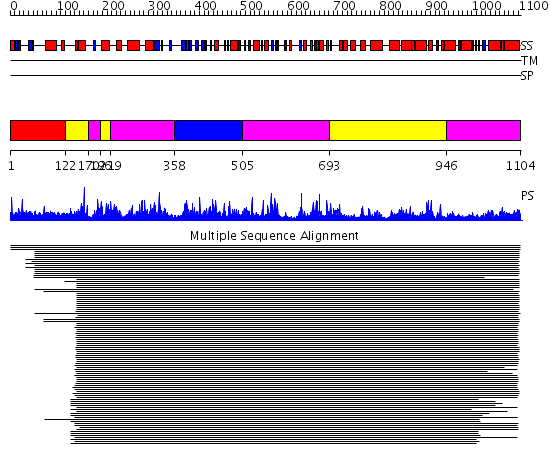
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 1.017968 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [122..169] [196..218] [693..945] |
2610.0 | Valyl-tRNA synthetase (ValRS) C-terminal domain; Valyl-tRNA synthetase (ValRS) | |
| 3 | View Details | [170..195] [219..357] [505..692] [946..1104] |
2610.0 | Valyl-tRNA synthetase (ValRS) C-terminal domain; Valyl-tRNA synthetase (ValRS) | |
| 4 | View Details | [358..504] | 2610.0 | Valyl-tRNA synthetase (ValRS) C-terminal domain; Valyl-tRNA synthetase (ValRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)