













| General Information: |
|
| Name(s) found: |
PRP22 /
YER013W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1145 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spliceosomal complex
[TAS |
| Biological Process: |
generation of catalytic spliceosome for second transesterification step
[IDA |
| Molecular Function: |
second spliceosomal transesterification activity
[IDA ATP-dependent RNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDISKLIGA IVGSDDPVII EFVLNIINKS GNLQEFIRNI QKLDAGISYE DSIKMYNAFL 60
61 GKQEEEKVRN KVKSSPLSQK INQVLKDDVN LDDPVVTEFV LSILNKSKSI TEFQEQLNLM 120
121 QSGLDNETIF KIYQIASPPV MKEEVSVLPS TKIPAKIEAK IEEEVQKIES LDPSPVLHKV 180
181 YEGKVRNITT FGCFVQIFGT RMKNCDGLVH ISEMSDQRTL DPHDVVRQGQ HIFVEVIKIQ 240
241 NNGKISLSMK NIDQHSGEIR KRNTESVEDR GRSNDAHTSR NMKNKIKRRA LTSPERWEIR 300
301 QLIASGAASI DDYPELKDEI PINTSYLTAK RDDGSIVNGN TEKVDSKLEE QQRDETDEID 360
361 VELNTDDGPK FLKDQQVKGA KKYEMPKITK VPRGFMNRSA INGSNAIRDH REEKLRKKRE 420
421 IEQQIRKQQS FDDPTKNKKD SRNEIQMLKN QLIVTEWEKN RMNESISYGK RTSLPISAQR 480
481 QTLPVYAMRS ELIQAVRDNQ FLVIVGETGS GKTTQITQYL DEEGFSNYGM IGCTQPRRVA 540
541 AVSVAKRVAE EVGCKVGHDV GYTIRFEDVT GPDTRIKYMT DGMLQREALL DPEMSKYSVI 600
601 MLDEAHERTV ATDVLFALLK KAAIKRPELK VIVTSATLNS AKFSEYFLNC PIINIPGKTF 660
661 PVEVLYSQTP QMDYIEAALD CVIDIHINEG PGDILVFLTG QEEIDSCCEI LYDRVKTLGD 720
721 SIGELLILPV YSALPSEIQS KIFEPTPKGS RKVVFATNIA ETSITIDGIY YVVDPGFAKI 780
781 NIYNARAGIE QLIVSPISQA QANQRKGRAG RTGPGKCYRL YTESAFYNEM LENTVPEIQR 840
841 QNLSHTILML KAMGINDLLK FDFMDPPPKN LMLNALTELY HLQSLDDEGK LTNLGKEMSL 900
901 FPMDPTLSRS LLSSVDNQCS DEIVTIISML SVQNVFYRPK DRQLEADSKK AKFHHPYGDH 960
961 LTLLNVYTRW QQANYSEQYC KTNFLHFRHL KRARDVKSQI SMIFKKIGLK LISCHSDPDL1020
1021 IRKTFVSGFF MNAAKRDSQV GYKTINGGTE VGIHPSSSLY GKEYEYVMYH SIVLTSREYM1080
1081 SQVTSIEPQW LLEVAPHFYK AGDAESQSRK KAKIIPLHNK FAKDQNSWRL SSIRQSRERA1140
1141 LGIKR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
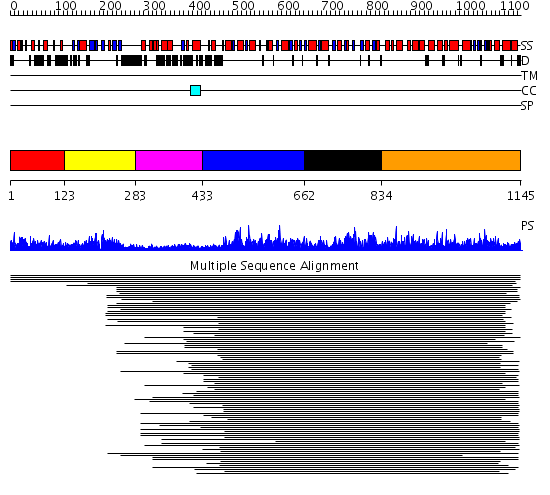
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 3.158997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [123..282] | 98.144602 | S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase | |
| 3 | View Details | [283..432] | 4.179995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [433..661] | 9.154902 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [662..833] | 9.154902 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 6 | View Details | [834..1145] | 12.325996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)