













| General Information: |
|
| Name(s) found: |
HSM3 /
YBR272C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 480 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
mismatch repair
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKETNYVE NLLTQLENEL NEDNLPEDIN TLLRKCSLNL VTVVSLPDMD VKPLLATIKR 60
61 FLTSNVSYDS LNYDYLLDVV DKLVPMADFD DVLEVYSAED LVKALRSEID PLKVAACRVI 120
121 ENSQPKGLFA TSNIIDILLD ILFDEKVEND KLITAIEKAL ERLSTDELIR RRLFDNNLPY 180
181 LVSVKGRMET VSFVRLIDFL TIEFQFISGP EFKDIIFCFT KEEILKSVED ILVFIELVNY 240
241 YTKFLLEIRN QDKYWALRHV KKILPVFAQL FEDTENYPDV RAFSTNCLLQ LFAEVSRIEE 300
301 DEYSLFKTMD KDSLKIGSEA KLITEWLELI NPQYLVKYHK DVVENYFHVS GYSIGMLRNL 360
361 SADEECFNAI RNKFSAEIVL RLPYLEQMQV VETLTRYEYT SKFLLNEMPK VMGSLIGDGS 420
421 AGAIIDLETV HYRNSALRNL LDKGEEKLSV WYEPLLREYS KAVNGKNYST GSETKIADCR 480
481 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |
|
HSM3 NAS6 RPN13 RPN14 RPT4 RVB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
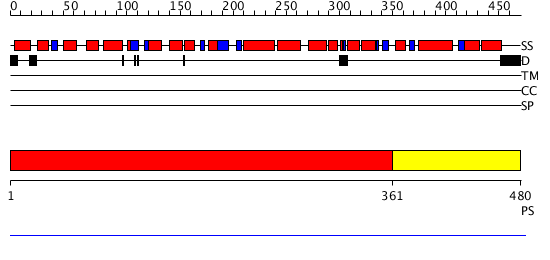
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..360] | 7.18 | No description for 1qoyA was found. | |
| 2 | View Details | [361..480] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)