













| General Information: |
|
| Name(s) found: |
GRS1 /
YBR121C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IMP mitochondrion [IDA |
| Biological Process: |
glycyl-tRNA aminoacylation
[IMP transcription termination [IMP |
| Molecular Function: |
glycine-tRNA ligase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVEDIKKAR AAVPFNREQL ESVLRGRFFY APAFDLYGGV SGLYDYGPPG CAFQNNIIDA 60
61 WRKHFILEED MLEVDCTMLT PYEVLKTSGH VDKFSDWMCR DLKTGEIFRA DHLVEEVLEA 120
121 RLKGDQEARG LVEDANAAAK DDAEKKKRKK KVKQIKAVKL DDDVVKEYEE ILAKIDGYSG 180
181 PELGELMEKY DIGNPVTGET LESPRAFNLM FETAIGPSGQ LKGYLRPETA QGQFLNFNKL 240
241 LEFNNSKTPF ASASIGKSFR NEISPRAGLL RVREFLMAEI EHFVDPLDKS HPKFNEIKDI 300
301 KLSFLPRDVQ EAGSTEPIVK TVGEAVASRM VDNETLGYFI ARIYQFLMKI GVDESKLRFR 360
361 QHMANEMAHY AADCWDGELK TSYGWIECVG CADRSAYDLT VHSKKTKEKL VVRQKLDNPI 420
421 EVTKWEIDLT KKLFGPKFRK DAPKVESHLL NMSQDDLASK AELLKANGKF TIKVDGVDGE 480
481 VELDDKLVKI EQRTKVEHVR EYVPSVIEPS FGIGRIIYSV FEHSFWNRPE DNARSVLSFP 540
541 PLVAPTKVLL VPLSNHKDLV PVHHEVAKIL RKSQIPFKID DSGVSIGKRY ARNDELGTPF 600
601 GVTIDFESAK DHSVTLRERD STKQVRGSVE NVIKAIRDIT YNGASWEEGT KDLTPFIAQA 660
661 EAEAETD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Hazbun TR, et al. (2003) |
|
GRS1 KAR2 TUB3 YNL313C |
| View Details | Ho Y, et al. (2002) |

|
AAT2 ACS2 ADE3 ADE6 ADH4 ADO1 AIM46 ALA1 APE2 ARA1 ARE1 ARG4 BAT1 CAR2 CDC10 CDC33 CDC60 CLU1 CPR1 CYS3 DED81 DPS1 ENP1 ERG10 ERG20 FET4 FUN12 GCY1 GPH1 GRS1 HAS1 HOM2 HYP2 ILS1 IMD2 IMD3 IMD4 KRE33 KRI1 KRS1 LSP1 MAG2 MDH3 MGM101 MPC54 MSC3 NAS6 NOC2 NOP12 NOP4 NPL3 OLA1 OYE2 PAB1 PDC5 PDC6 PFK2 PGM2 PMI40 PRP6 RPF2 RRP12 SAC6 SCP160 SOD1 SSZ1 SUI1 THR4 TIF4631 TIF4632 TPS1 TRR1 UBA1 UTP22 YDL124W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
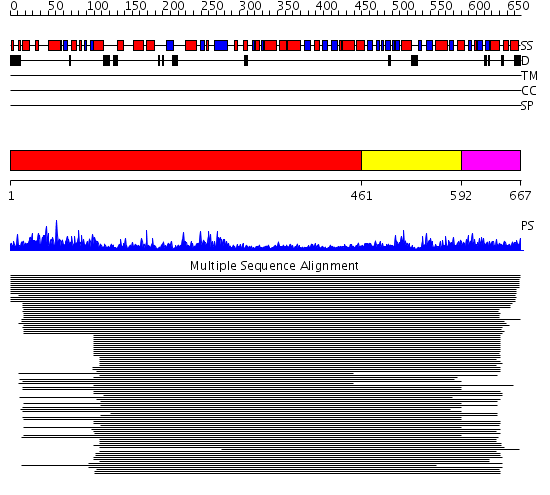
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..460] | 437.457575 | Glycyl-tRNA synthetase (GlyRS), C-terminal domain; Glycyl-tRNA synthetase (GlyRS) | |
| 2 | View Details | [461..591] | 437.457575 | Glycyl-tRNA synthetase (GlyRS), C-terminal domain; Glycyl-tRNA synthetase (GlyRS) | |
| 3 | View Details | [592..667] | 51.045757 | Threonyl-tRNA synthetase (ThrRS), C-terminal domain; Threonyl-tRNA synthetase (ThrRS), N-terminal 'additional' domain; Threonyl-tRNA synthetase (ThrRS), second 'additional' domain; Threonyl-tRNA synthetase (ThrRS) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)