













| General Information: |
|
| Name(s) found: |
UBP15 /
YMR304W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1230 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC |
| Biological Process: |
protein deubiquitination
[TAS |
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSEDELGSI GTVFPGSPID KSIGSILPQF DEEVETLLED SFTWNIPDWN ELTNPKYNSP 60
61 RFRIGDFEWD ILLFPQGNHN KGVAVYLEPH PEEKLDETTG EMVPVDPDWY CCAQFAIGIS 120
121 RPGNGDTINL INKSHHRFNA LDTDWGFANL IDLNNLKHPS KGRPLSFLNE GTLNITAYVR 180
181 ILKDPTGVLW HNFLNYDSKK VTGYVGFRNQ GATCYLNSLL QSYFFTKYFR KLVYEIPTEH 240
241 ESPNNSVPLA LQRAFYQLQV SDIPLDTLEL TRSFGWDTAE SFTQHDVQEL NRILMDRLEN 300
301 NMKGTPVEGK LNEIFVGKMK SYIKCINVDY ESARVEDFWD LQLNVKNFKN LQESFDNYIE 360
361 MELMNGENQY AAQDYGLQDA QKGVIFESFP PVLHLQLKRF EYDFNYDQMV KVNDKYEFPE 420
421 TIDLSPFVDK DVLKKTLDSE NKDKNPYVYN LHGVLVHSGD ISTGHYYTLI KPGVEDQWYR 480
481 FDDERVWRVT KKQVFQENFG CDRLPDEKVR TMTRGEYQNY IIQRHTSAYM LVYIRQEQEE 540
541 DLLRPVLESD VPKHVITRVR EEIKERETKE KEIREAHLYV TLRLHSIKEF IHYEGFDYFA 600
601 HDGFRLFAEE LNDSGLQQIN LKVLRTTKLS DIFASIKETM NIPQERDVKY WKMDYRRNST 660
661 LRLTQPINFE SVNITLQEAL KKEKKRTMQT QYGEEGVAST EEDDKALLET VSFLDLFIEE 720
721 PYLELQFLNK LKEASLISKA QLDDELISTI RTNLPELTKG GIEPVFATDN KSNLLFVKSY 780
781 DPHTQKLLGF GHFAVNQLQQ LSDISAIIED SISSNEKLTF YEEVQPGTIN EIYMKETIYD 840
841 ADIDTGDIVS FEVPGAVLPD TFPVYATIKD FYSYLRYRVK LKFSKFDGSS EEYGVSNEIP 900
901 ESFEFWISAY APYDDLARMV SKYAHVKPEY LKIIALYSNG RFVLKSTSLL NDYLLKDFNC 960
961 DQIPPFAFEV LSVPLKELER LRPIKLYWLK NSYIHYQCFE FEVANDYTES QFLEKVQHKI1020
1021 GFTDEEKENI LLWTNTNFQF QGLLSDQNTF KDVSKHSLLF GRILPEESKL FKELNRLENV1080
1081 QTSSLEDFMD DENATDRPMD DEQDLGMAIE HSEDMKGRIV VVQQYFKDLE NRHGISFLFN1140
1141 LIPDETFPKT KDRLHAKFGL GQKEFSKIKL SIGYSTEEGT VFRSLQGFSD EELDKVILYD1200
1201 IMSNLDYIYM DHPDRLRSHS SYDRPMIIKN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ECM30 UBP15 |
| View Details | Krogan NJ, et al. (2006) |
|
ECM30 PAD1 RRT1 UBP15 |
| View Details | Gavin AC, et al. (2006) |

|
ECM30 UBP15 |
| View Details | Gavin AC, et al. (2002) |
|
ACE2 ALT2 BCK1 CDC11 DRE2 ECM30 ENP2 IML1 MNL1 MTR4 MYO2 RGC1 SEC3 UBP15 YBL044W YJR098C YNL313C |
| View Details | Ho Y, et al. (2002) |

|
ADK1 AHC1 AHP1 ARO1 CBK1 CCT2 CCT3 CDC28 CDC39 CDH1 CKB2 CLB2 CLU1 COP1 CUE5 DCS1 DCS2 DNM1 ECM10 FAR1 GAL7 GDB1 GPH1 HSL7 KEL1 LSB3 LSM4 NAN1 NAP1 PAT1 PRB1 PRP11 RAD50 REX2 RPT3 SEC26 SEC27 SGT2 SHM2 SRP1 SSD1 SSK2 SWE1 TFP1 THI22 TIF4631 TPS1 TSA2 UBP15 UBR1 UTP13 VPS1 YAK1 YFR024C YGR250C YJL045W YLR241W YLR422W YME2 YPL247C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
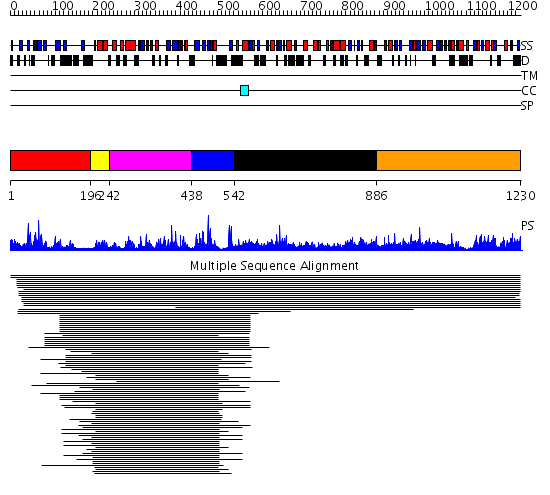
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | 15.30103 | MATH domain No confident structure predictions are available. | |
| 2 | View Details | [196..241] | 12.60206 | No description for PF00442 was found. No confident structure predictions are available. | |
| 3 | View Details | [242..437] | 5.245991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [438..541] | 30.187087 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. | |
| 5 | View Details | [542..885] | 1.061952 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [886..1230] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)