













| General Information: |
|
| Name(s) found: |
YLR422W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1932 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQQDSQRWL PTDRLIYGVL VKSFLPLQRY PELVYENSNY ANVYVGAEVY VFEESVDKKW 60
61 CRAYQCLRPF PEEFISNMNS ANDVLPDVKP KVVIFPRKYV HFEAEKAVST MPFFKAPSAE 120
121 DFKPLISKEC ESRSFCDSLY VSSTDDISTG KPRKTPRPPF PFFRYQKRSF KDEMGPILSL 180
181 ISSHVYSMYS IGEFSIYRKM IKLYYDLDTI RFRLSMNLTT EAEKINLIRA ATSLRTKIAK 240
241 FLSSTYRKNK LIANSTPRNP DPYGFEGIFA RDIDTGELLS YEIDKLRTLV SSSMLCGLTN 300
301 NFPTVPVVES DDESSSNGLF GTVRSSILVN LKDLAWDPSI SDPKYQDLSI CVYLRTKDEV 360
361 LTESFTMTKS SNMESALDEI PAMLFKNILE TIVHKNKVYL VVVLKETIAI TTETAPEISS 420
421 YNISTEESSS HSPFSPFNSS TENKIDHVKK GLAAGVINIS PVFKFYNGLS VANKAQRFNL 480
481 YLYSSDSSDS QNFNSSKDAD LGWGGLINKI IKDSSEGVSV NPRAVSLSVT VKEIIGKQEA 540
541 EKVLSTSLVP IRSIPTYFYD TMFSQAERIY LNLGRVSLYG LPAADTNIEN VTVQISCRNK 600
601 AVKFCKNKLE ERSGDWKFVS VRPNESIGES IRIEGVENMN EDETLRVLVY LNGFLMAKSN 660
661 IHIKKKNEII EYRKGTVFQI MSSKSVPLIH LELEASYFGR RYNINPAITN FLVLQTKNVE 720
721 FDQQLKEHYS VTLKQLNNVS FKDLLKHFDT ILAHYLLLLE SVNEATDKKG PSSSLPNIVF 780
781 SEFVKFLNLM LTHQENSRYW FNRLYKKVMS KELECPNVAP ILIKHMTTIF DRSHSSWTRT 840
841 GTAICRTILY IIVLAIGSSH SDEMPNFSHF FRSLHKFLML ADEPIMADQI LLIESIPSML 900
901 ETMTNHCKVE DLVRFAIGLF ECCQEKEMNQ KMYSRPLSVR EEEYLNTKFN CLLKLINKKV 960
961 LQNYLTNTES VDKLRLQFLS KTLEWLLTPY TPGDDKCFHV ESLRLVNSVF ITIIEDYKFD1020
1021 MLQRNLIRLL PYLCKSFVHL RRYCKKARLM RPRRVFTMLF PREIPCNYIP VDSIVNDEVV1080
1081 VEVLLELAII ICEITKIASS RFPSYQSFSE IINLCDKDTL FQSNFYSRQI TNENVYTITK1140
1141 TVFLFFKQDW FPGMKWLGVS ALLGRSSLIL LSLCKDYIIE NNSPSPSKES EKRVDMRLWA1200
1201 EYVKVILLVS NHKSASLTKL AITPRKAVYL ISGDLKKISA YILNECWDAL ATGHYNITYA1260
1261 KKYGLGALSD CQFELFVHNQ FLIREIFIFA FHRHIDATRI CCKILWGLGL NFWRIFGSLQ1320
1321 PAVNACIPEL FSAYQIGKLR LNDYELERFV SCLFFMMHVP DSDTFFPACM DFLRDLLGFL1380
1381 HIVNEIYKIP NQEEFDDDRT ARHIEMFEYL LEANRPELFH KMIYDLFIHF IQKKDFVQAA1440
1441 LSLELLAGTY AWDSNDTLEA ISFPPLPEQS SFERKEYLLK ESARNFSRGQ KPEKALAVYK1500
1501 DLIKAYDEIN YDLNGLAFVH DQIAGIYTRL QSIDRLVPTY FKVSFMGFGF PKSLRNKSFV1560
1561 FEGLPFEHIT SMHDRLLRSY HGSNIVHSQE EVDMLLMNPP MGKYIHVASV EPCLSISDNY1620
1621 NSSDKKSSIN NKVRMYIENR DLRTFSNSRR LPGAKGVTDL WVEEYTYHTM NTFPTLMNRS1680
1681 EIVKVTKSKL SPLENAIRSL QVKIQELYGL ENMCNKTLKD HGDVNDLFTE LSTNITGTIS1740
1741 APVNGGISQY KAFLEPSTSK QFSTDDLGRL TLAFDELVAV LGRCLTLHAE LLPSKDLKPS1800
1801 HDLLVRLFEE NFAEEIERYS RTLSEANRSR NNMITARIIS HKNPNKKASF SGRDHHTSGS1860
1861 NHSQFVLEHS DSFGPNSLLF GKYLTRTLSH SSTTSSLDKS GIVSGTSSTF LAGSQPNTNT1920
1921 DSQHKHDYSH SG |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
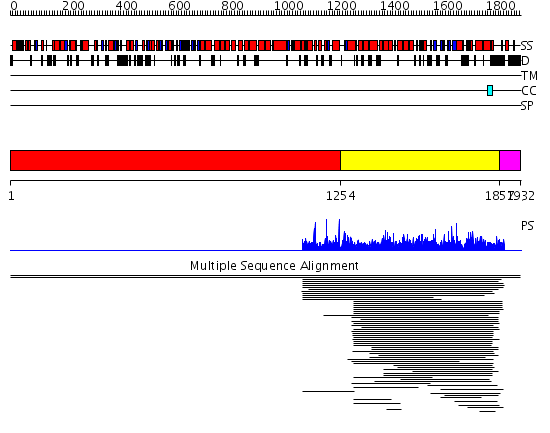
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1253] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [1254..1856] | 9.037877 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1857..1932] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [435..498] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [499..586] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [587..770] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [771..987] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [988..1101] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1102..1165] | 2.065996 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1166..1269] | 1.068988 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1270..1307] | 1.091996 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1308..1395] | 2.106986 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1396..1468] | 1.125982 | View MSA. No confident structure predictions are available. | |
| 14 | View Details | [1469..1533] | 2.127994 | View MSA. No confident structure predictions are available. | |
| 15 | View Details | [1534..1626] | 2.131985 | View MSA. No confident structure predictions are available. | |
| 16 | View Details | [1627..1691] | N/A | No confident structure predictions are available. | |
| 17 | View Details | [1692..1846] | 8.131981 | View MSA. No confident structure predictions are available. | |
| 18 | View Details | [1847..1932] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. | |||||||||
| 10 | No functions predicted. | |||||||||
| 11 | No functions predicted. | |||||||||
| 12 | No functions predicted. | |||||||||
| 13 | No functions predicted. | |||||||||
| 14 | No functions predicted. | |||||||||
| 15 | No functions predicted. | |||||||||
| 16 | No functions predicted. | |||||||||
| 17 | No functions predicted. | |||||||||
| 18 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)