













| General Information: |
|
| Name(s) found: |
ALA1 /
YOR335C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 958 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
alanyl-tRNA aminoacylation
[IGI |
| Molecular Function: |
alanine-tRNA ligase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIGDKQKWT ATNVRNTFLD YFKSKEHKFV KSSPVVPFDD PTLLFANAGM NQYKPIFLGT 60
61 VDPASDFYTL KRAYNSQKCI RAGGKHNDLE DVGKDSYHHT FFEMLGNWSF GDYFKKEAIT 120
121 YSWTLLTEVY GIPKDRLYVT YFEGDEKLGL EPDTEARELW KNVGVPDDHI LPGNAKDNFW 180
181 EMGDQGPCGP CSEIHYDRIG GRNAASLVNM DDPDVLEVWN LVFIQFNREQ DGSLKPLPAK 240
241 HIDTGMGFER LVSVLQDVRS NYDTDVFTPL FERIQEITSV RPYSGNFGEN DKDGIDTAYR 300
301 VLADHVRTLT FALADGGVPN NEGRGYVLRR ILRRGARYAR KYMNYPIGNF FSTLAPTLIS 360
361 QVQDIFPELA KDPAFLFEIL DEEEASFAKT LDRGERLFEK YASAASKTES KTLDGKQVWR 420
421 LYDTYGFPVD LTELMAEEQG LKIDGPGFEK AKQESYEASK RGGKKDQSDL IKLNVHELSE 480
481 LNDAKVPKTN DEFKYGSANV EGTILKLHDG TNFVDEITEP GKKYGIILDK TCFYAEQGGQ 540
541 EYDTGKIVID DAAEFNVENV QLYNGFVFHT GSLEEGKLSV GDKIIASFDE LRRFPIKNNH 600
601 TGTHILNFAL KETLGNDVDQ KGSLVAPEKL RFDFSHKKAV SNEELKKVED ICNEQIKENL 660
661 QVFYKEIPLD LAKSIDGVRA VFGETYPDPV RVVSVGKPIE ELLANPANEE WTKYSIEFCG 720
721 GTHVNKTGDI KYFVILEESG IAKGIRRIVA VTGTEAFEAQ RLAEQFAADL DAADKLPFSP 780
781 IKEKKLKELG VKLGQLSISV ITKNELKQKF NKIEKAVKDE VKSRAKKENK QTLDEVKTFF 840
841 ETNENAPYLV KFIDISPNAK AITEAINYMK SNDSVKDKSI YLLAGNDPEG RVAHGCYISN 900
901 AALAKGIDGS ALAKKVSSII GGKAGGKGNV FQGMGDKPAA IKDAVDDLES LFKEKLSI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
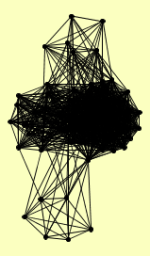
| View Details | Krogan NJ, et al. (2006) |

|
ALA1 ARO1 DUG1 GDH1 HIS4 HIS6 HRD1 MET6 OYE2 RAD14 SCP160 SPE3 SPE4 TIM11 URA6 YLF2 YRB2 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACS2 ADE3 ADE6 ADH4 ADO1 ALA1 APE2 ARA1 ARG4 AXL1 BAT1 CAR2 CDC10 CDC60 CLU1 CPR1 CYS3 DBF20 DED81 DPS1 EGD2 ENP1 ERG10 ERG20 FPR1 GCY1 GPH1 GRS1 HOM2 HOM3 HYP2 IDH2 ILS1 IMD3 KRS1 LSP1 MDH3 MLP2 MOB1 MPC54 NAS6 OLA1 PAB1 PDC5 PFK2 PGM2 PMI40 PRP6 RPB10 SAC6 SCP160 SOD1 SSZ1 SUI1 THR4 THS1 TPS1 TRR1 UBA1 URA1 YDL124W YDR341C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
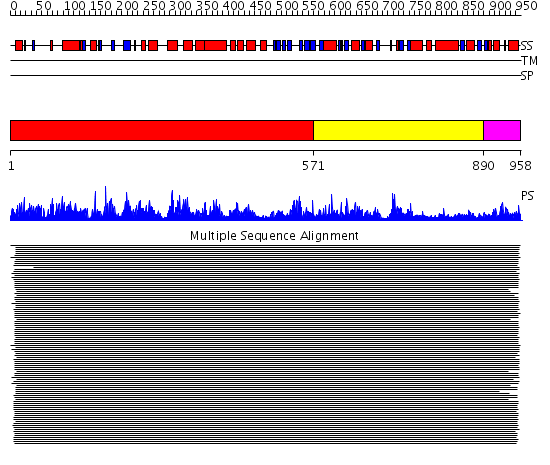
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..570] | 288.026872 | tRNA synthetases class II (A) No confident structure predictions are available. | |
| 2 | View Details | [571..889] | 65.69897 | Threonyl-tRNA synthetase (ThrRS), C-terminal domain; Threonyl-tRNA synthetase (ThrRS), N-terminal 'additional' domain; Threonyl-tRNA synthetase (ThrRS), second 'additional' domain; Threonyl-tRNA synthetase (ThrRS) | |
| 3 | View Details | [890..958] | 4.182996 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [876..958] | 8.240996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)