













| General Information: |
|
| Name(s) found: |
HSH155 /
YMR288W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 971 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U2 snRNP
[IDA |
| Biological Process: |
spliceosome assembly
[IPI |
| Molecular Function: |
mRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHPIQFVNA NNSDKSHQLG GQYSIPQDLR ENLQKEAARI GENEKDVLQE KMETRTVQNR 60
61 EDSYHKRRFD MKFEPDSDTQ TVTSSENTQD AVVPRKRKSR WDVKGYEPPD ESSTAVKENS 120
121 DSALVNVEGI HDLMFFKPSD HKYFADVISK KPIDELNKDE KKERTLSMLL LKIKNGNTAS 180
181 RRTSMRILTD KAVTFGPEMI FNRLLPILLD RSLEDQERHL MIKTIDRVLY QLGDLTKPYV 240
241 HKILVVAAPL LIDEDPMVRS TGQEIITNLS TVAGLKTILT VMRPDIENED EYVRNVTSRA 300
301 AAVVAKALGV NQLLPFINAA CHSRKSWKAR HTGIKIVQQI GILLGIGVLN HLTGLMSCIK 360
361 DCLMDDHVPV RIVTAHTLST LAENSYPYGI EVFNVVLEPL WKGIRSHRGK VLSSFLKAVG 420
421 SMIPLMDPEY AGYYTTEAMR IIRREFDSPD DEMKKTILLV LQKCSAVESI TPKFLREEIA 480
481 PEFFQKFWVR RVALDRPLNK VVTYTTVTLA KKLGCSYTID KLLTPLRDEA EPFRTMAVHA 540
541 VTRTVNLLGT ADLDERLETR LIDALLIAFQ EQTNSDSIIF KGFGAVTVSL DIRMKPFLAP 600
601 IVSTILNHLK HKTPLVRQHA ADLCAILIPV IKNCHEFEML NKLNIILYES LGEVYPEVLG 660
661 SIINAMYCIT SVMDLDKLQP PINQILPTLT PILRNKHRKV EVNTIKFVGL IGKLAPTYAP 720
721 PKEWMRICFE LLELLKSTNK EIRRSANATF GFIAEAIGPH DVLVALLNNL KVQERQLRVC 780
781 TAVAIGIVAK VCGPYNVLPV IMNEYTTPET NVQNGVLKAM SFMFEYIGNM SKDYIYFITP 840
841 LLEDALTDRD LVHRQTASNV ITHLALNCSG TGHEDAFIHL MNLLIPNIFE TSPHAIMRIL 900
901 EGLEALSQAL GPGLFMNYIW AGLFHPAKNV RKAFWRVYNN MYVMYQDAMV PFYPVTPDNN 960
961 EEYIEELDLV L |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
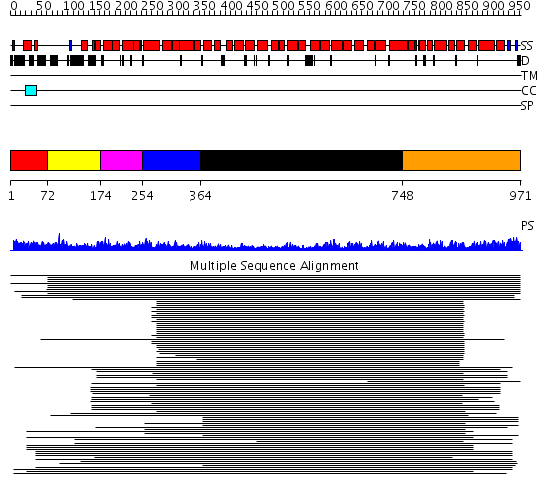
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [72..173] | 1.152985 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [174..253] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [254..363] | 82.39794 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [364..747] | 55.0 | Importin beta | |
| 6 | View Details | [748..971] | 55.0 | Importin beta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)