













| General Information: |
|
| Name(s) found: |
MSH6 /
YDR097C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1242 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
meiotic mismatch repair
[IMP mismatch repair [IDA |
| Molecular Function: |
ATPase activity
[IDA ATP binding [IDA four-way junction DNA binding [IDA guanine/thymine mispair binding [IDA single base insertion or deletion binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPATPKTSK TAHFENGSTS SQKKMKQSSL LSFFSKQVPS GTPSKKVQKP TPATLENTAT 60
61 DKITKNPQGG KTGKLFVDVD EDNDLTIAEE TVSTVRSDIM HSQEPQSDTM LNSNTTEPKS 120
121 TTTDEDLSSS QSRRNHKRRV NYAESDDDDS DTTFTAKRKK GKVVDSESDE DEYLPDKNDG 180
181 DEDDDIADDK EDIKGELAED SGDDDDLISL AETTSKKKFS YNTSHSSSPF TRNISRDNSK 240
241 KKSRPNQAPS RSYNPSHSQP SATSKSSKFN KQNEERYQWL VDERDAQRRP KSDPEYDPRT 300
301 LYIPSSAWNK FTPFEKQYWE IKSKMWDCIV FFKKGKFFEL YEKDALLANA LFDLKIAGGG 360
361 RANMQLAGIP EMSFEYWAAQ FIQMGYKVAK VDQRESMLAK EMREGSKGIV KRELQCILTS 420
421 GTLTDGDMLH SDLATFCLAI REEPGNFYNE TQLDSSTIVQ KLNTKIFGAA FIDTATGELQ 480
481 MLEFEDDSEC TKLDTLMSQV RPMEVVMERN NLSTLANKIV KFNSAPNAIF NEVKAGEEFY 540
541 DCDKTYAEII SSEYFSTEED WPEVLKSYYD TGKKVGFSAF GGLLYYLKWL KLDKNLISMK 600
601 NIKEYDFVKS QHSMVLDGIT LQNLEIFSNS FDGSDKGTLF KLFNRAITPM GKRMMKKWLM 660
661 HPLLRKNDIE SRLDSVDSLL QDITLREQLE ITFSKLPDLE RMLARIHSRT IKVKDFEKVI 720
721 TAFETIIELQ DSLKNNDLKG DVSKYISSFP EGLVEAVKSW TNAFERQKAI NENIIVPQRG 780
781 FDIEFDKSMD RIQELEDELM EILMTYRKQF KCSNIQYKDS GKEIYTIEIP ISATKNVPSN 840
841 WVQMAANKTY KRYYSDEVRA LARSMAEAKE IHKTLEEDLK NRLCQKFDAH YNTIWMPTIQ 900
901 AISNIDCLLA ITRTSEYLGA PSCRPTIVDE VDSKTNTQLN GFLKFKSLRH PCFNLGATTA 960
961 KDFIPNDIEL GKEQPRLGLL TGANAAGKST ILRMACIAVI MAQMGCYVPC ESAVLTPIDR1020
1021 IMTRLGANDN IMQGKSTFFV ELAETKKILD MATNRSLLVV DELGRGGSSS DGFAIAESVL1080
1081 HHVATHIQSL GFFATHYGTL ASSFKHHPQV RPLKMSILVD EATRNVTFLY KMLEGQSEGS1140
1141 FGMHVASMCG ISKEIIDNAQ IAADNLEHTS RLVKERDLAA NNLNGEVVSV PGGLQSDFVR1200
1201 IAYGDGLKNT KLGSGEGVLN YDWNIKRNVL KSLFSIIDDL QS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
MPH1 MSH2 MSH6 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
MSH2 MSH6 |
| View Details | Krogan NJ, et al. (2006) |
|
MSH2 MSH6 |
| View Details | Gavin AC, et al. (2006) |
|
MPH1 MSH6 RAD52 RFA1 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 ECM29 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MRPS28 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 RSC8 RVB2 SGS1 SHS1 SRO7 SSL1 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |

|
MSH2 MSH6 |
| View Details | Qiu et al. (2008) |
|
MSH2 MSH3 MSH6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
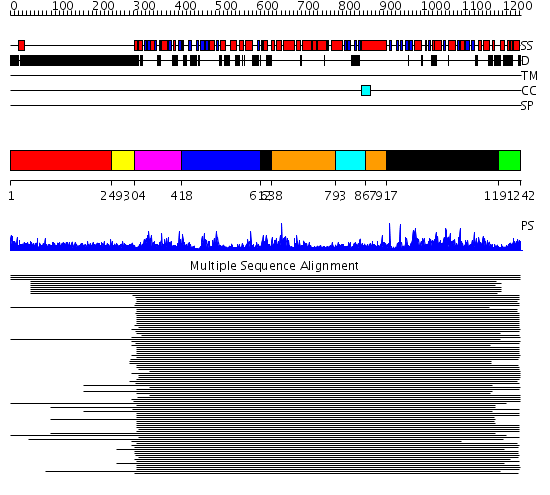
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 10.28 | No description for 1ldvA was found. | |
| 2 | View Details | [249..303] | 8.11 | RBP1 | |
| 3 | View Details | [304..417] | 1615.228787 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 4 | View Details | [418..611] | 1615.228787 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 5 | View Details | [612..637] [917..1190] |
1615.228787 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 6 | View Details | [638..792] [867..916] |
1615.228787 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 7 | View Details | [793..866] | 1615.228787 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 8 | View Details | [1191..1242] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)