













| General Information: |
|
| Name(s) found: |
DUR1,2 /
YBR208C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1835 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
urea metabolic process
[IMP allantoin catabolic process [TAS |
| Molecular Function: |
allophanate hydrolase activity
[IMP urea carboxylase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVSSDTTAE ISLGWSIQDW IDFHKSSSSQ ASLRLLESLL DSQNVAPVDN AWISLISKEN 60
61 LLHQFQILKS RENKETLPLY GVPIAVKDNI DVRGLPTTAA CPSFAYEPSK DSKVVELLRN 120
121 AGAIIVGKTN LDQFATGLVG TRSPYGKTPC AFSKEHVSGG SSAGSASVVA RGIVPIALGT 180
181 DTAGSGRVPA ALNNLIGLKP TKGVFSCQGV VPACKSLDCV SIFALNLSDA ERCFRIMCQP 240
241 DPDNDEYSRP YVSNPLKKFS SNVTIAIPKN IPWYGETKNP VLFSNAVENL SRTGANVIEI 300
301 DFEPLLELAR CLYEGTWVAE RYQAIQSFLD SKPPKESLDP TVISIIEGAK KYSAVDCFSF 360
361 EYKRQGILQK VRRLLESVDV LCVPTCPLNP TMQQVADEPV LVNSRQGTWT NFVNLADLAA 420
421 LAVPAGFRDD GLPNGITLIG KKFTDYALLE LANRYFQNIF PNGSRTYGTF TSSSVKPAND 480
481 QLVGPDYDPS TSIKLAVVGA HLKGLPLHWQ LEKVNATYLC TTKTSKAYQL FALPKNGPVL 540
541 KPGLRRVQDS NGSQIELEVY SVPKELFGAF ISMVPEPLGI GSVELESGEW IKSFICEESG 600
601 YKAKGTVDIT KYGGFRAYFE MLKKKESQKK KLFDTVLIAN RGEIAVRIIK TLKKLGIRSV 660
661 AVYSDPDKYS QHVTDADVSV PLHGTTAAQT YLDMNKIIDA AKQTNAQAII PGYGFLSENA 720
721 DFSDACTSAG ITFVGPSGDI IRGLGLKHSA RQIAQKAGVP LVPGSLLITS VEEAKKVAAE 780
781 LEYPVMVKST AGGGGIGLQK VDSEEDIEHI FETVKHQGET FFGDAGVFLE RFIENARHVE 840
841 VQLMGDGFGK AIALGERDCS LQRRNQKVIE ETPAPNLPEK TRLALRKAAE SLGSLLNYKC 900
901 AGTVEFIYDE KKDEFYFLEV NTRLQVEHPI TEMVTGLDLV EWMIRIAAND APDFDSTKVE 960
961 VNGVSMEARL YAENPLKNFR PSPGLLVDVK FPDWARVDTW VKKGTNISPE YDPTLAKIIV1020
1021 HGKDRDDAIS KLNQALEETK VYGCITNIDY LKSIITSDFF AKAKVSTNIL NSYQYEPTAI1080
1081 EITLPGAHTS IQDYPGRVGY WRIGVPPSGP MDAYSFRLAN RIVGNDYRTP AIEVTLTGPS1140
1141 IVFHCETVIA ITGGTALCTL DGQEIPQHKP VEVKRGSTLS IGKLTSGCRA YLGIRGGIDV1200
1201 PKYLGSYSTF TLGNVGGYNG RVLKLGDVLF LPSNEENKSV ECLPQNIPQS LIPQISETKE1260
1261 WRIGVTCGPH GSPDFFKPES IEEFFSEKWK VHYNSNRFGV RLIGPKPKWA RSNGGEGGMH1320
1321 PSNTHDYVYS LGAINFTGDE PVIITCDGPS LGGFVCQAVV PEAELWKVGQ VKPGDSIQFV1380
1381 PLSYESSRSL KESQDVAIKS LDGTKLRRLD SVSILPSFET PILAQMEKVN ELSPKVVYRQ1440
1441 AGDRYVLVEY GDNEMNFNIS YRIECLISLV KKNKTIGIVE MSQGVRSVLI EFDGYKVTQK1500
1501 ELLKVLVAYE TEIQFDENWK ITSNIIRLPM AFEDSKTLAC VQRYQETIRS SAPWLPNNVD1560
1561 FIANVNGISR NEVYDMLYSA RFMVLGLGDV FLGSPCAVPL DPRHRFLGSK YNPSRTYTER1620
1621 GAVGIGGMYM CIYAANSPGG YQLVGRTIPI WDKLCLAASS EVPWLMNPFD QVEFYPVSEE1680
1681 DLDKMTEDCD NGVYKVNIEK SVFDHQEYLR WINANKDSIT AFQEGQLGER AEEFAKLIQN1740
1741 ANSELKESVT VKPDEEEDFP EGAEIVYSEY SGRFWKSIAS VGDVIEAGQG LLIIEAMKAE1800
1801 MIISAPKSGK IIKICHGNGD MVDSGDIVAV IETLA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
DOA4 DUR1,2 MRPL22 PRO1 |
| View Details | Gavin AC, et al. (2002) |
|
APM1 CYM1 DOA4 DUR1,2 HSP42 LAT1 LPD1 PDA1 PDB1 PDX1 PRP46 RPN11 TCM62 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 BBC1 BNI1 CDC39 CDC53 CRM1 DUR1,2 ECM29 ECM33 FAA4 GAL3 GCN1 HRT1 HYP2 IMH1 KAP122 MCM4 MCM7 MDN1 MET18 MKT1 MMS1 MYO2 PFK1 PMA2 RPA190 RPN1 RPT4 SAP185 SIT4 TIP41 TPS1 TRP3 UBI4 VPS13 YAR009C YGP1 YJR029W YLR035C-A YMR209C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
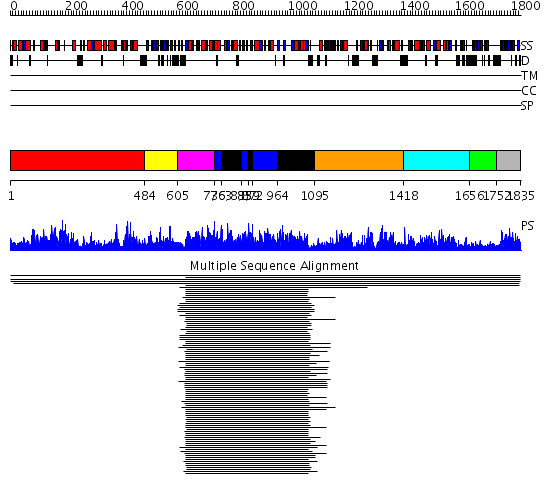
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..483] | 194.980454 | No description for 1gr8A_ was found. | |
| 2 | View Details | [484..604] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [605..735] | 1882.218487 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 4 | View Details | [736..762] [833..858] [872..963] |
1882.218487 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 5 | View Details | [763..832] [859..871] [964..1094] |
1882.218487 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 6 | View Details | [1095..1417] | 190.244125 | Allophanate hydrolase subunit 2 No confident structure predictions are available. | |
| 7 | View Details | [1418..1655] | 3.008774 | Allophanate hydrolase subunit 1 No confident structure predictions are available. | |
| 8 | View Details | [1656..1751] | N/A | Confident ab initio structure predictions are available. | |
| 9 | View Details | [1752..1835] | 46.09691 | Biotin carboxyl carrier domain of transcarboxylase (TC 1.3S) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)