













| General Information: |
|
| Name(s) found: |
YDJ1 /
YNL064C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 409 amino acids |
Gene Ontology: |
|
| Cellular Component: |
perinuclear region of cytoplasm
[IDA microsome [IDA cytosol [IDA |
| Biological Process: |
protein targeting to mitochondrion
[IMP response to drug [IMP protein targeting to ER [IMP ER-associated protein catabolic process [IGI protein refolding [IDA 'de novo' protein folding [IMP |
| Molecular Function: |
Hsp70/Hsc70 protein regulator activity
[IDA unfolded protein binding [IDA chaperone regulator activity [IGI ATPase activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKETKFYDI LGVPVTATDV EIKKAYRKCA LKYHPDKNPS EEAAEKFKEA SAAYEILSDP 60
61 EKRDIYDQFG EDGLSGAGGA GGFPGGGFGF GDDIFSQFFG AGGAQRPRGP QRGKDIKHEI 120
121 SASLEELYKG RTAKLALNKQ ILCKECEGRG GKKGAVKKCT SCNGQGIKFV TRQMGPMIQR 180
181 FQTECDVCHG TGDIIDPKDR CKSCNGKKVE NERKILEVHV EPGMKDGQRI VFKGEADQAP 240
241 DVIPGDVVFI VSERPHKSFK RDGDDLVYEA EIDLLTAIAG GEFALEHVSG DWLKVGIVPG 300
301 EVIAPGMRKV IEGKGMPIPK YGGYGNLIIK FTIKFPENHF TSEENLKKLE EILPPRIVPA 360
361 IPKKATVDEC VLADFDPAKY NRTRASRGGA NYDSDEEEQG GEGVQCASQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
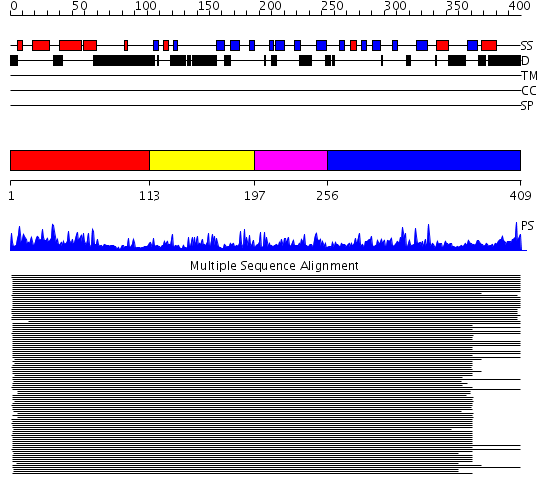
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 193.228787 | HSP40 | |
| 2 | View Details | [113..196] | 18.0 | Cysteine-rich domain of the chaperone protein DnaJ. | |
| 3 | View Details | [197..255] | 146.315398 | Heat shock protein 40 Sis1 | |
| 4 | View Details | [256..409] | 146.315398 | Heat shock protein 40 Sis1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)