













| General Information: |
|
| Name(s) found: |
STE11 /
YLR362W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
activation of MAPKK activity
[IDA MAPKKK cascade involved in osmosensory signaling pathway [IGI pseudohyphal growth [IMP osmosensory signaling pathway via Sho1 osmosensor [IGI invasive growth in response to glucose limitation [IMP signal transduction during filamentous growth [IMP protein amino acid phosphorylation [IDA pheromone-dependent signal transduction involved in conjugation with cellular fusion [IMP |
| Molecular Function: |
SAM domain binding
[IDA MAP kinase kinase kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQTQTAEGT DLLIGDEKTN DLPFVQLFLE EIGCTQYLDS FIQCNLVTEE EIKYLDKDIL 60
61 IALGVNKIGD RLKILRKSKS FQRDKRIEQV NRLKNLMEKV SSLSTATLSM NSELIPEKHC 120
121 VIFILNDGSA KKVNVNGCFN ADSIKKRLIR RLPHELLATN SNGEVTKMVQ DYDVFVLDYT 180
181 KNVLHLLYDV ELVTICHAND RVEKNRLIFV SKDQTPSDKA ISTSKKLYLR TLSALSQVGP 240
241 SSSNLLAQNK GISHNNAEGK LRIDNTEKDR IRQIFNQRPP SEFISTNLAG YFPHTDMKRL 300
301 QKTMRESFRH SARLSIAQRR PLSAESNNIG DILLKHSNAV DMALLQGLDQ TRLSSKLDTT 360
361 KIPKLAHKRP EDNDAISNQL ELLSVESGEE EDHDFFGEDS DIVSLPTKIA TPKNWLKGAC 420
421 IGSGSFGSVY LGMNAHTGEL MAVKQVEIKN NNIGVPTDNN KQANSDENNE QEEQQEKIED 480
481 VGAVSHPKTN QNIHRKMVDA LQHEMNLLKE LHHENIVTYY GASQEGGNLN IFLEYVPGGS 540
541 VSSMLNNYGP FEESLITNFT RQILIGVAYL HKKNIIHRDI KGANILIDIK GCVKITDFGI 600
601 SKKLSPLNKK QNKRASLQGS VFWMSPEVVK QTATTAKADI WSTGCVVIEM FTGKHPFPDF 660
661 SQMQAIFKIG TNTTPEIPSW ATSEGKNFLR KAFELDYQYR PSALELLQHP WLDAHII |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
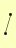
|
STE11 STE50 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
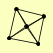
|
FUS3 KSS1 STE11 STE5 STE7 |
| View Details | Krogan NJ, et al. (2006) |

|
STE11 STE50 |
| View Details | Gavin AC, et al. (2006) |

|
STE11 STE50 |
| View Details | Ho Y, et al. (2002) |
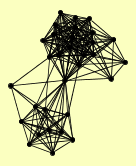
|
AVO1 BCK2 BEM3 CCT2 CYS4 DIG1 DIG2 GDI1 GUS1 KSS1 NPA3 PIM1 RNH203 RPA135 RPN6 RVB1 SEC4 STE11 STE12 STE7 TEC1 TSC11 VPS21 YPT1 YPT10 YPT31 YPT32 YPT52 YPT53 YPT6 YPT7 |
| View Details | Qiu et al. (2008) |
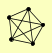
|
FUS3 KSS1 STE11 STE5 STE7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
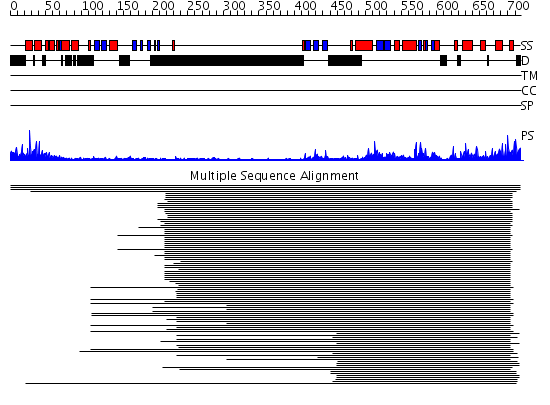
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 8.30103 | NMR structure of the Saccharomyces cerevisiae SAM (Sterile Alpha Motif) domain | |
| 2 | View Details | [107..210] | 2.33 | Protein kinase byr2 | |
| 3 | View Details | [211..717] | 66.39794 | No description for 2h8hA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)