













| General Information: |
|
| Name(s) found: |
GPA1 /
YHR005C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 472 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA heterotrimeric G-protein complex [IDA plasma membrane [IDA |
| Biological Process: |
response to drug
[IMP heterotrimeric G-protein complex cycle [IMP inositol lipid-mediated signaling [IMP pheromone-dependent signal transduction involved in conjugation with cellular fusion [IMP |
| Molecular Function: |
GTPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGCTVSTQTI GDESDPFLQN KRANDVIEQS LQLEKQRDKN EIKLLLLGAG ESGKSTVLKQ 60
61 LKLLHQGGFS HQERLQYAQV IWADAIQSMK ILIIQARKLG IQLDCDDPIN NKDLFACKRI 120
121 LLKAKALDYI NASVAGGSDF LNDYVLKYSE RYETRRRVQS TGRAKAAFDE DGNISNVKSD 180
181 TDRDAETVTQ NEDADRNNSS RINLQDICKD LNQEGDDQMF VRKTSREIQG QNRRNLIHED 240
241 IAKAIKQLWN NDKGIKQCFA RSNEFQLEGS AAYYFDNIEK FASPNYVCTD EDILKGRIKT 300
301 TGITETEFNI GSSKFKVLDA GGQRSERKKW IHCFEGITAV LFVLAMSEYD QMLFEDERVN 360
361 RMHESIMLFD TLLNSKWFKD TPFILFLNKI DLFEEKVKSM PIRKYFPDYQ GRVGDAEAGL 420
421 KYFEKIFLSL NKTNKPIYVK RTCATDTQTM KFVLSAVTDL IIQQNLKKIG II |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GPA1 STE18 STE4 |
| View Details | Ho Y, et al. (2002) |
|
AHA1 ARP2 CCT2 CCT3 CCT5 CCT6 GCD11 GPA1 GPA2 RNQ1 STE4 TCP1 YHR033W |
| View Details | Qiu et al. (2008) |
|
GPA1 STE18 STE4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
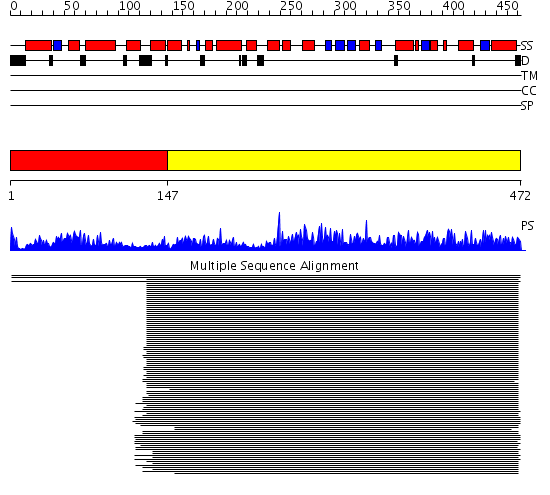
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 5.823909 | G-protein alpha subunit No confident structure predictions are available. | |
| 2 | View Details | [147..472] | 1559.0 | No description for 1scgA_ was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)