













| General Information: |
|
| Name(s) found: |
VTC3 /
YPL019C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 835 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar membrane
[IDA |
| Biological Process: |
vacuole fusion, non-autophagic
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLFGIKLAND VYPPWKDSYI DYERLKKLLK ESVIHDGRSS VDSWSERNES DFVEALDKEL 60
61 EKVYTFQISK YNAVLRKLDD LEENTKSAEK IQKINSEQFK NTLEECLDEA QRLDNFDRLN 120
121 FTGFIKIVKK HDKLHPNYPS VKSLLQVRLK ELPFNNSEEY SPLLYRISYL YEFLRSNYDH 180
181 PNTVSKSLAS TSKLSHFSNL EDASFKSYKF WVHDDNIMEV KARILRHLPA LVYASVPNEN 240
241 DDFVDNLESD VRVQPEARLN IGSKSNSLSS DGNSNQDVEI GKSKSVIFPQ SYDPTITTLY 300
301 FDNDFFDLYN NRLLKISGAP TLRLRWIGKL LDKPDIFLEK RTFTENTETG NSSFEEIRLQ 360
361 MKAKFINNFI FKNDPSYKNY LINQLRERGT QKEELEKLSR DFDNIQNFIV EEKLQPVLRA 420
421 TYNRTAFQIP GDQSIRVTID SNIMYIREDS LDKNRPIRNP ENWHRDDIDS NIPNPLRFLR 480
481 AGEYSKFPYS VMEIKVINQD NSQMPNYEWI KDLTNSHLVN EVPKFSLYLQ GVASLFGEDD 540
541 KYVNILPFWL PDLETDIRKN PQEAYEEEKK TLQKQKSIHD KLDNMRRLSK ISVPDGKTTE 600
601 RQGQKDQNTR HVIADLEDHE SSDEEGTALP KKSAVKKGKK FKTNAAFLKI LAGKNISENG 660
661 NDPYSDDTDS ASSFQLPPGV KKPVHLLKNA GPVKVEAKVW LANERTFNRW LSVTTLLSVL 720
721 TFSIYNSVQK AEFPQLADLL AYVYFFLTLF CGVWAYRTYL KRLTLIKGRS GKHLDAPVGP 780
781 ILVAVVLIVT LVVNFSVAFK EAARRERGLV NVSSQPSLPR TLKPIQDFIF NLVGE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
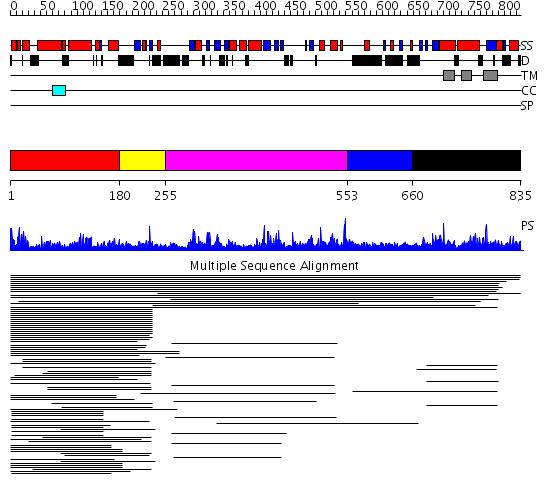
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 53.443697 | SPX domain No confident structure predictions are available. | |
| 2 | View Details | [180..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..552] | 4.027969 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [553..659] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [660..835] | 4.021997 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|