













| General Information: |
|
| Name(s) found: |
MCM7 /
YBR202W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 845 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IDA cytoplasm [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[TAS pre-replicative complex assembly [IPI DNA unwinding involved in replication [TAS |
| Molecular Function: |
ATP binding
[IDA chromatin binding [TAS ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAALPSIQL PVDYNNLFNE ITDFLVTFKQ DTLSSDATRN ENEDENLDAE NIEQHLLEKG 60
61 PKYMAMLQKV ANRELNSVII DLDDILQYQN EKFLQGTQAD DLVSAIQQNA NHFTELFCRA 120
121 IDNNMPLPTK EIDYKDDVLD VILNQRRLRN ERMLSDRTNE IRSENLMDTT MDPPSSMNDA 180
181 LREVVEDETE LFPPNLTRRY FLYFKPLSQN CARRYRKKAI SSKPLSVRQI KGDFLGQLIT 240
241 VRGIITRVSD VKPAVEVIAY TCDQCGYEVF QEVNSRTFTP LSECTSEECS QNQTKGQLFM 300
301 STRASKFSAF QECKIQELSQ QVPVGHIPRS LNIHVNGTLV RSLSPGDIVD VTGIFLPAPY 360
361 TGFKALKAGL LTETYLEAQF VRQHKKKFAS FSLTSDVEER VMELITSGDV YNRLAKSIAP 420
421 EIYGNLDVKK ALLLLLVGGV DKRVGDGMKI RGDINVCLMG DPGVAKSQLL KAICKISPRG 480
481 VYTTGKGSSG VGLTAAVMKD PVTDEMILEG GALVLADNGI CCIDEFDKMD ESDRTAIHEV 540
541 MEQQTISISK AVINTNPGAR TSILAAANPL YGRINPRLSP LDNINLPAAL LSRFDILFLM 600
601 LDIPSRDDDE KLAEHVTYVH MHNKQPDLDF TPVEPSKMRE YIAYAKTKRP VMSEAVNDYV 660
661 VQAYIRLRQD SKREMDSKFS FGQATPRTLL GIIRLSQALA KLRLADMVDI DDVEEALRLV 720
721 RVSKESLYQE TNKSKEDESP TTKIFTIIKK MLQETGKNTL SYENIVKTVR LRGFTMLQLS 780
781 NCIQEYSYLN VWHLINEGNT LKFVDDGTMD TDQEDSLVST PKLAPQTTAS ANVSAQDSDI 840
841 DLQDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CDC45 CDC6 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 SLD3 |
| View Details | Gavin AC, et al. (2006) |
|
ERG26 MCM7 MYO5 |
| View Details | Gavin AC, et al. (2002) |

|
CDC45 ERG26 GLT1 HAT1 HSP42 LYS12 MCM2 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 RPD3 RPN9 SIN3 TFP1 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 BNI1 DUR1,2 IMH1 KAP122 MCM4 MCM7 MET18 RPT4 TRP3 YJR029W |
| View Details | Qiu et al. (2008) |
|
CDC45 CDC6 DPB11 DPB2 DPB3 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 RFC1 RFC4 RRM3 SLD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
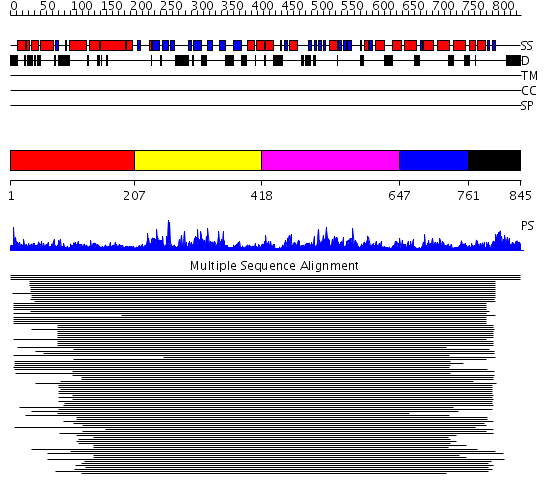
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | 1.089991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [207..417] | 8.79588 | MCM2/3/5 family No confident structure predictions are available. | |
| 3 | View Details | [418..646] | 46.522879 | ATPase subunit of magnesium chelatase, BchI | |
| 4 | View Details | [647..760] | 46.522879 | ATPase subunit of magnesium chelatase, BchI | |
| 5 | View Details | [761..845] | 2.270992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)