













| General Information: |
|
| Name(s) found: |
MOT1 /
YPL082C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1867 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA nuclear chromosome [IDA nuclear outer membrane [IMP |
| Biological Process: |
transcription of nuclear rRNA large RNA polymerase I transcript
[IDA regulation of RNA polymerase II transcriptional preinitiation complex assembly [IMP regulation of transcription from RNA polymerase II promoter [IDA |
| Molecular Function: |
ATPase activity
[IDA rDNA binding [IDA TATA-binding protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSRVSRLDR QVILIETGST QVVRNMAADQ MGDLAKQHPE DILSLLSRVY PFLLVKKWET 60
61 RVTAARAVGG IVAHAPSWDP NESDLVGGTN EGSPLDNAQV KLEHEMKIKL EEATQNNQLN 120
121 LLQEDHHLSS LSDWKLNEIL KSGKVLLASS MNDYNVLGKA DDNIRKQAKT DDIKQETSML 180
181 NASDKANENK SNANKKSARM LAMARRKKKM SAKNTPKHPV DITESSVSKT LLNGKNMTNS 240
241 AASLATSPTS NQLNPKLEIT EQADESKLMI ESTVRPLLEQ HEIVAGLVWQ FQGIYELLLD 300
301 NLMSENWEIR HGAALGLREL VKKHAYGVSR VKGNTREENN LRNSRSLEDL ASRLLTVFAL 360
361 DRFGDYVYDT VVAPVRESVA QTLAALLIHL DSTLSIKIFN CLEQLVLQDP LQTGLPNKIW 420
421 EATHGGLLGI RYFVSIKTNF LFAHGLLENV VRIVLYGLNQ SDDDVQSVAA SILTPITSEF 480
481 VKLNNSTIEI LVTTIWSLLA RLDDDISSSV GSIMDLLAKL CDHQEVLDIL KNKALEHPSE 540
541 WSFKSLVPKL YPFLRHSISS VRRAVLNLLI AFLSIKDDST KNWLNGKVFR LVFQNILLEQ 600
601 NPELLQLSFD VYVALLEHYK VKHTEKTLDH VFSKHLQPIL HLLNTPVGEK GKNYAMESQY 660
661 ILKPSQHYQL HPEKKRSISE TTTDSDIPIP KNNEHINIDA PMIAGDITLL GLDVILNTRI 720
721 MGAKAFALTL SMFQDSTLQS FFTNVLVRCL ELPFSTPRML AGIIVSQFCS SWLQKHPEGE 780
781 KLPSFVSEIF SPVMNKQLLN RDEFPVFREL VPSLKALRTQ CQSLLATFVD VGMLPQYKLP 840
841 NVAIVVQGET EAGPHAFGVE TAEKVYGEYY DKMFKSMNNS YKLLAKKPLE DSKHRVLMAI 900
901 NSAKESAKLR TGSILANYAS SILLFDGLPL KLNPIIRSLM DSVKEERNEK LQTMAGESVV 960
961 HLIQQLLENN KVNVSGKIVK NLCGFLCVDT SEVPDFSVNA EYKEKILTLI KESNSIAAQD1020
1021 DINLAKMSEE AQLKRKGGLI TLKILFEVLG PSILQKLPQL RSILFDSLSD HENEEASKVD1080
1081 NEQGQKIVDS FGVLRALFPF MSDSLRSSEV FTRFPVLLTF LRSNLSVFRY SAARTFADLA1140
1141 KISSVEVMAY TIREILPLMN SAGSLSDRQG STELIYHLSL SMETDVLPYV IFLIVPLLGR1200
1201 MSDSNEDVRN LATTTFASII KLVPLEAGIA DPKGLPEELV ASRERERDFI QQMMDPSKAK1260
1261 PFKLPIAIKA TLRKYQQDGV NWLAFLNKYH LHGILCDDMG LGKTLQTICI IASDQYLRKE1320
1321 DYEKTRSVES RALPSLIICP PSLTGHWENE FDQYAPFLKV VVYAGGPTVR LTLRPQLSDA1380
1381 DIIVTSYDVA RNDLAVLNKT EYNYCVLDEG HIIKNSQSKL AKAVKEITAN HRLILTGTPI1440
1441 QNNVLELWSL FDFLMPGFLG TEKMFQERFA KPIAASRNSK TSSKEQEAGV LALEALHKQV1500
1501 LPFMLRRLKE DVLSDLPPKI IQDYYCELGD LQKQLYMDFT KKQKNVVEKD IENSEIADGK1560
1561 QHIFQALQYM RKLCNHPALV LSPNHPQLAQ VQDYLKQTGL DLHDIINAPK LSALRTLLFE1620
1621 CGIGEEDIDK KASQDQNFPI QNVISQHRAL IFCQLKDMLD MVENDLFKKY MPSVTYMRLD1680
1681 GSIDPRDRQK VVRKFNEDPS IDCLLLTTKV GGLGLNLTGA DTVIFVEHDW NPMNDLQAMD1740
1741 RAHRIGQKKV VNVYRIITKG TLEEKIMGLQ KFKMNIASTV VNQQNSGLAS MDTHQLLDLF1800
1801 DPDNVTSQDN EEKNNGDSQA AKGMEDIANE TGLTGKAKEA LGELKELWDP SQYEEEYNLD1860
1861 TFIKTLR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
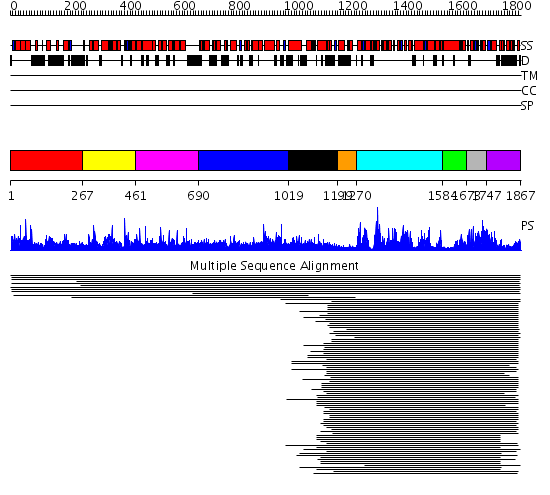
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..266] | 1.011923 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [267..460] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [461..689] | 1.01494 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [690..1018] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1019..1198] | 2.326994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1199..1269] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1270..1583] | 151.040959 | SNF2 family N-terminal domain No confident structure predictions are available. | |
| 8 | View Details | [1584..1672] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1673..1746] | 24.638272 | Helicase conserved C-terminal domain No confident structure predictions are available. | |
| 10 | View Details | [1747..1867] | 7.580996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)