













| General Information: |
|
| Name(s) found: |
MSB1 /
YOR188W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1137 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud tip
[TAS cellular bud neck [TAS mitochondrion [IDA |
| Biological Process: |
cell division
[RCA]
regulation of transport [RCA] establishment of cell polarity [IGI] regulation of signal transduction [RCA] reproduction [RCA] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDMAKPLPT PPTAEIRKSR SNSPKKAQKT NLSPNKNQNN EKNVPRSNGR TKNEHNSMDD 60
61 EEFEFFHQFS REKVKGVIHV ITAELKEKGP DVEFLMIPFR PEQTNDKLLT LLNQLFPLGN 120
121 GQPVNEKKQL RIVSKADVWT LFQCLKYIWC RLPNSEIIGW KSYLEFKFRE EDKKFPRKSF 180
181 LEIMPQCLAS PNHASIVYDF FDLIISISSN SRVNKMSARK ISKMCAIWAF SKQIPNSDIQ 240
241 DYDFESAAMK SFAPNNSIQD GLDQWIPASD AMFHLLLAFL RSFVPQDLES AKLPRTLKSL 300
301 LFNNQYPPRK STAYTSETIL TIPLVTLKTD VFSRKPWQLL ERCNDLLDFS DHDAFEARED 360
361 YALLKSLFRK KNTVEGISRK MSQESRRLMK AMSTKHSTFQ PGWAPRECIE NISHLKECIE 420
421 VKRLDIDDYF IWTWLSSLSF EQTSEKKKIF GRSIILEFEF DGFKKWVVFQ ECDITLDYNK 480
481 KGQLKKKTSA QSPTTEKELP PDDFELEDPP LSKSPTLSQT YKKFQAEVPQ QSTVRRDSAP 540
541 DNQGIYHTVI SKNALTKNKH NVNLHSFEHK ISKWNPLNNL RKKSGSNSSS SSFEEKSKDA 600
601 PIREEYHTNK NHKSKKEERV LSQFSTLNPD EYQLPVIETG SSNFKIEIPE LMYEHDDDDS 660
661 DKLKNSQKRA TDSAIEELNG MVEEMMINEP DDVKISITEA ETFESLTKFD QYKPSNITDD 720
721 DLQSSHSSAV HSLKLSTNTN DSCADSSKYT ADRKLAEPRK ISEESKVNDD SSSYYSPNIN 780
781 NLPASRMPSQ PTYSNSDSKK AFTNESRLNV LQGAVSPSQQ VTPKPYKNAP GDCVSPVQQK 840
841 YYQNDRRNEM SPASAPVPPS AYSPARSPQF STNSAGFKQN TINVPVGYND PAHVLANQPH 900
901 MTYRDQHNYP SHQQKQRPFQ NNIVPPELKS RNQRADASPI PQHMVPVKQG VPNLPSNVPL 960
961 YQQMERMNPN HQHPVNTYKV TQPPYHNNTT NAYGNSRAGN AHMLDGKWSN NPPQMVPKGV1020
1021 RPNQYPQQHV NRYSPQAQPV VPAEYYNGPP PMRAPPMMSH MVPAQEPIRY TAGANRRSFP1080
1081 QGMQQNAYSV PAQPMGAVNS EFYLPEAPQG NKLHGNINKR QERKKLYDNI RSGNFGI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
MSB1 RIM8 YPL216W |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Qiu et al. (2008) |

|
BEM1 MSB1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
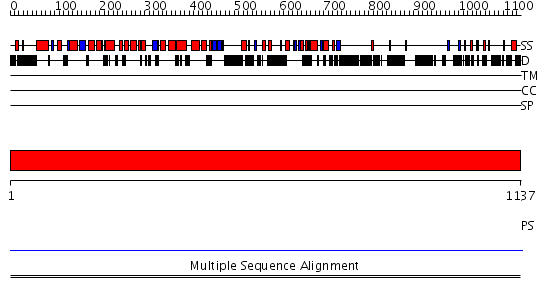
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1137] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [309..588] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [589..771] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [772..929] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [930..1137] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)