













| General Information: |
|
| Name(s) found: |
SPT20 /
YOL148C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 604 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SLIK (SAGA-like) complex
[IPI SAGA complex [IDA |
| Biological Process: |
chromatin modification
[TAS histone acetylation [TAS |
| Molecular Function: |
transcription cofactor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSANSPTGND PHVFGIPVNA TPSNMGSPGS PVNVPPPMNP AVANVNHPVM RTNSNSNANE 60
61 GTRTLTREQI QQLQQRQRLL LQQRLLEQQR KQQALQNYEA QFYQMLMTLN KRPKRLYNFV 120
121 EDADSILKKY EQYLHSFEFH IYENNYKICA PANSRLQQQQ KQPELTSDGL ILTKNNETLK 180
181 EFLEYVARGR IPDAIMEVLR DCNIQFYEGN LILQVYDHTN TVDVTPKENK PNLNSSSSPS 240
241 NNNSTQDNSK IQQPSEPNSG VANTGANTAN KKASFKRPRV YRTLLKPNDL TTYYDMMSYA 300
301 DNARFSDSIY QQFESEILTL TKRNLSLSVP LNPYEHRDML EETAFSEPHW DSEKKSFIHE 360
361 HRAESTREGT KGVVGHIEER DEFPQHSSNY EQLMLIMNER TTTITNSTFA VSLTKNAMEI 420
421 ASSSSNGVRG ASSSTSNSAS NTRNNSLANG NQVALAAAAA AAAVGSTMGN DNNQFSRLKF 480
481 IEQWRINKEK RKQQALSANI NPTPFNARIS MTAPLTPQQQ LLQRQQQALE QQQNGGAMKN 540
541 ANKRSGNNAT SNNNNNNNNL DKPKVKRPRK NAKKSESGTP APKKKRMTKK KQSASSTPSS 600
601 TTMS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
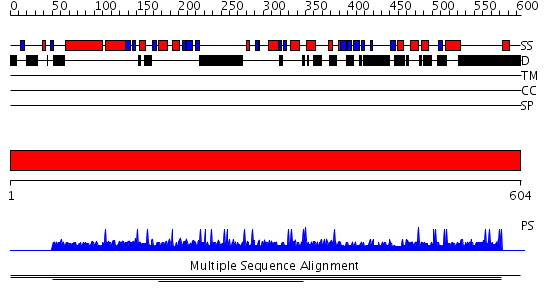
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..604] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)