













| General Information: |
|
| Name(s) found: |
REV3 /
YPL167C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA zeta DNA polymerase complex [IDA |
| Biological Process: |
mutagenesis
[IGI DNA repair [TAS |
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] DNA binding [IEA] DNA-directed DNA polymerase activity [IDA][IEA] nucleotidyltransferase activity [IEA] metal ion binding [IEA] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRESNDTIQ SDTVRSSSKS DYFRIQLNNQ DYYMSKPTFL DPSHGESLPL NQFSQVPNIR 60
61 VFGALPTGHQ VLCHVHGILP YMFIKYDGQI TDTSTLRHQR CAQVHKTLEV KIRASFKRKK 120
121 DDKHDLAGDK LGNLNFVADV SVVKGIPFYG YHVGWNLFYK ISLLNPSCLS RISELIRDGK 180
181 IFGKKFEIYE SHIPYLLQWT ADFNLFGCSW INVDRCYFRS PVLNSILDID KLTINDDLQL 240
241 LLDRFCDFKC NVLSRRDFPR VGNGLIEIDI LPQFIKNREK LQHRDIHHDF LEKLGDISDI 300
301 PVKPYVSSAR DMINELTMQR EELSLKEYKE PPETKRHVSG HQWQSSGEFE AFYKKAQHKT 360
361 STFDGQIPNF ENFIDKNQKF SAINTPYEAL PQLWPRLPQI EINNNSMQDK KNDDQVNASF 420
421 TEYEICGVDN ENEGVKGSNI KSRSYSWLPE SIASPKDSTI LLDHQTKYHN TINFSMDCAM 480
481 TQNMASKRKL RSSVSANKTS LLSRKRKKVM AAGLRYGKRA FVYGEPPFGY QDILNKLEDE 540
541 GFPKIDYKDP FFSNPVDLEN KPYAYAGKRF EISSTHVSTR IPVQFGGETV SVYNKPTFDM 600
601 FSSWKYALKP PTYDAVQKWY NKVPSMGNKK TESQISMHTP HSKFLYKFAS DVSGKQKRKK 660
661 SSVHDSLTHL TLEIHANTRS DKIPDPAIDE VSMIIWCLEE ETFPLDLDIA YEGIMIVHKA 720
721 SEDSTFPTKI QHCINEIPVM FYESEFEMFE ALTDLVLLLD PDILSGFEIH NFSWGYIIER 780
781 CQKIHQFDIV RELARVKCQI KTKLSDTWGY AHSSGIMITG RHMINIWRAL RSDVNLTQYT 840
841 IESAAFNILH KRLPHFSFES LTNMWNAKKS TTELKTVLNY WLSRAQINIQ LLRKQDYIAR 900
901 NIEQARLIGI DFHSVYYRGS QFKVESFLIR ICKSESFILL SPGKKDVRKQ KALECVPLVM 960
961 EPESAFYKSP LIVLDFQSLY PSIMIGYNYC YSTMIGRVRE INLTENNLGV SKFSLPRNIL1020
1021 ALLKNDVTIA PNGVVYAKTS VRKSTLSKML TDILDVRVMI KKTMNEIGDD NTTLKRLLNN1080
1081 KQLALKLLAN VTYGYTSASF SGRMPCSDLA DSIVQTGRET LEKAIDIIEK DETWNAKVVY1140
1141 GDTDSLFVYL PGKTAIEAFS IGHAMAERVT QNNPKPIFLK FEKVYHPSIL ISKKRYVGFS1200
1201 YESPSQTLPI FDAKGIETVR RDGIPAQQKI IEKCIRLLFQ TKDLSKIKKY LQNEFFKIQI1260
1261 GKVSAQDFCF AKEVKLGAYK SEKTAPAGAV VVKRRINEDH RAEPQYKERI PYLVVKGKQG1320
1321 QLLRERCVSP EEFLEGENLE LDSEYYINKI LIPPLDRLFN LIGINVGNWA QEIVKSKRAS1380
1381 TTTTKVENIT RVGTSATCCN CGEELTKICS LQLCDDCLEK RSTTTLSFLI KKLKRQKEYQ1440
1441 TLKTVCRTCS YRYTSDAGIE NDHIASKCNS YDCPVFYSRV KAERYLRDNQ SVQREEALIS1500
1501 LNDW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
REV3 REV7 |
| View Details | Krogan NJ, et al. (2006) |

|
ADA2 AHC1 AHC2 FMP40 GCN5 HFI1 MSS4 NGG1 PDR15 RAD59 REV3 SGF11 SGF29 SGF73 SNL1 SPT20 SPT3 SPT7 SPT8 TAF1 TAF10 TAF11 TAF12 TAF13 TAF3 TAF4 TAF5 TAF6 TAF7 TAF9 UBP8 YMR007W |
| View Details | Qiu et al. (2008) |
|
DPB3 POL3 POL31 POL32 REV3 REV7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
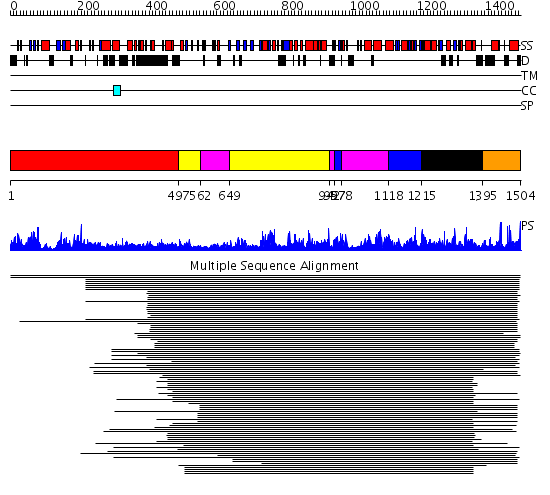
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..496] | 1.135927 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [497..561] [649..941] |
1346.9897 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 3 | View Details | [562..648] [942..956] [978..1117] |
1346.9897 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 4 | View Details | [957..977] [1118..1214] |
1346.9897 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 5 | View Details | [1215..1394] | 1346.9897 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 6 | View Details | [1395..1504] | 1.096987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)