













| General Information: |
|
| Name(s) found: |
MTR4 /
YJL050W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1073 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [TAS nucleolus [IDA TRAMP complex [IDA |
| Biological Process: |
mRNA export from nucleus
[IMP rRNA catabolic process [IMP ribosome biogenesis [RCA tRNA catabolic process [IDA ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process [IDA polyadenylation-dependent ncRNA catabolic process [IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSTDLFDVF EETPVELPTD SNGEKNADTN VGDTPDHTQD KKHGLEEEKE EHEENNSENK 60
61 KIKSNKSKTE DKNKKVVVPV LADSFEQEAS REVDASKGLT NSETLQVEQD GKVRLSHQVR 120
121 HQVALPPNYD YTPIAEHKRV NEARTYPFTL DPFQDTAISC IDRGESVLVS AHTSAGKTVV 180
181 AEYAIAQSLK NKQRVIYTSP IKALSNQKYR ELLAEFGDVG LMTGDITINP DAGCLVMTTE 240
241 ILRSMLYRGS EVMREVAWVI FDEVHYMRDK ERGVVWEETI ILLPDKVRYV FLSATIPNAM 300
301 EFAEWICKIH SQPCHIVYTN FRPTPLQHYL FPAHGDGIYL VVDEKSTFRE ENFQKAMASI 360
361 SNQIGDDPNS TDSRGKKGQT YKGGSAKGDA KGDIYKIVKM IWKKKYNPVI VFSFSKRDCE 420
421 ELALKMSKLD FNSDDEKEAL TKIFNNAIAL LPETDRELPQ IKHILPLLRR GIGIHHSGLL 480
481 PILKEVIEIL FQEGFLKVLF ATETFSIGLN MPAKTVVFTS VRKWDGQQFR WVSGGEYIQM 540
541 SGRAGRRGLD DRGIVIMMID EKMEPQVAKG MVKGQADRLD SAFHLGYNMI LNLMRVEGIS 600
601 PEFMLEHSFF QFQNVISVPV MEKKLAELKK DFDGIEVEDE ENVKEYHEIE QAIKGYREDV 660
661 RQVVTHPANA LSFLQPGRLV EISVNGKDNY GWGAVVDFAK RINKRNPSAV YTDHESYIVN 720
721 VVVNTMYIDS PVNLLKPFNP TLPEGIRPAE EGEKSICAVI PITLDSIKSI GNLRLYMPKD 780
781 IRASGQKETV GKSLREVNRR FPDGIPVLDP VKNMKIEDED FLKLMKKIDV LNTKLSSNPL 840
841 TNSMRLEELY GKYSRKHDLH EDMKQLKRKI SESQAVIQLD DLRRRKRVLR RLGFCTPNDI 900
901 IELKGRVACE ISSGDELLLT ELIFNGNFNE LKPEQAAALL SCFAFQERCK EAPRLKPELA 960
961 EPLKAMREIA AKIAKIMKDS KIEVVEKDYV ESFRHELMEV VYEWCRGATF TQICKMTDVY1020
1021 EGSLIRMFKR LEELVKELVD VANTIGNSSL KEKMEAVLKL IHRDIVSAGS LYL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
AIR1 AIR2 MTR4 NAB3 PAP2 PRP43 |
| View Details | Krogan NJ, et al. (2006) |
|
AIR1 AIR2 MTR4 PAP2 PSO2 RPL18A, RPL18B STE4 TRF5 |
| View Details | Gavin AC, et al. (2006) |
|
MTR4 NAB3 NRD1 SEN1 STO1 |
| View Details | Gavin AC, et al. (2002) |
|
ACE2 ALT2 BCK1 CDC11 DRE2 ECM30 ENP2 GCN1 IML1 MNL1 MTR4 MYO2 NAB3 RGC1 SEC3 UBP15 YBL044W YJR098C YNL313C |
| View Details | Ho Y, et al. (2002) |
|
AIR1 AIR2 IMD1 IMD3 IMD4 MTR4 NAP1 NOP53 NOP56 PAP2 PSE1 |
| View Details | Qiu et al. (2008) |
|
AIR1 AIR2 DBP10 DBP3 DBP6 DBP7 DBP8 DHR2 DRS1 FAL1 GAR1 HAS1 HCA4 MAK5 MTR4 NSR1 PAP2 POP1 RNT1 RRP3 SPB4 SUB2 UTP13 UTP18 UTP4 UTP6 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample 3121 run on DecaLCQ: searched for phosphorylation | Green EM, et al (2005) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
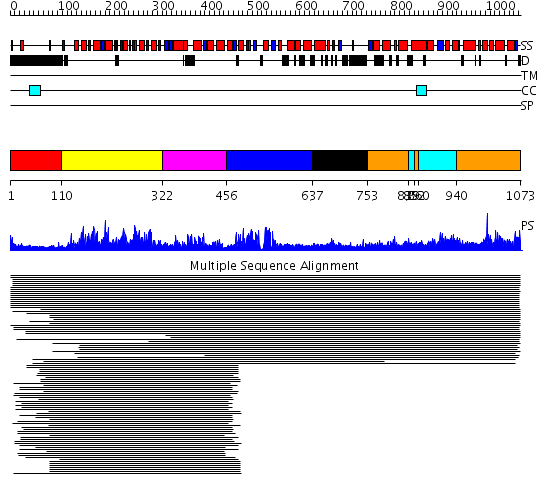
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..321] | 89.522879 | Putative DEAD box RNA helicase | |
| 3 | View Details | [322..455] | 89.522879 | Putative DEAD box RNA helicase | |
| 4 | View Details | [456..636] | 25.39794 | Initiation factor 4a | |
| 5 | View Details | [637..752] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [753..838] [852..859] [940..1073] |
8.05 | (+)-bornyl diphosphate synthase | |
| 7 | View Details | [839..851] [860..939] |
8.05 | (+)-bornyl diphosphate synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)