













| General Information: |
|
| Name(s) found: |
PPT1 /
YGR123C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 513 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
protein amino acid dephosphorylation
[IDA ribosome biogenesis [RCA |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTPTAADRA KALERKNEGN VFVKEKHFLK AIEKYTEAID LDSTQSIYFS NRAFAHFKVD 60
61 NFQSALNDCD EAIKLDPKNI KAYHRRALSC MALLEFKKAR KDLNVLLKAK PNDPAATKAL 120
121 LTCDRFIREE RFRKAIGGAE NEAKISLCQT LNLSSFDANA DLANYEGPKL EFEQLYDDKN 180
181 AFKGAKIKNM SQEFISKMVN DLFLKGKYLP KKYVAAIISH ADTLFRQEPS MVELENNSTP 240
241 DVKISVCGDT HGQFYDVLNL FRKFGKVGPK HTYLFNGDFV DRGSWSCEVA LLFYCLKILH 300
301 PNNFFLNRGN HESDNMNKIY GFEDECKYKY SQRIFNMFAQ SFESLPLATL INNDYLVMHG 360
361 GLPSDPSATL SDFKNIDRFA QPPRDGAFME LLWADPQEAN GMGPSQRGLG HAFGPDITDR 420
421 FLRNNKLRKI FRSHELRMGG VQFEQKGKLM TVFSAPNYCD SQGNLGGVIH VVPGHGILQA 480
481 GRNDDQNLII ETFEAVEHPD IKPMAYSNGG FGL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
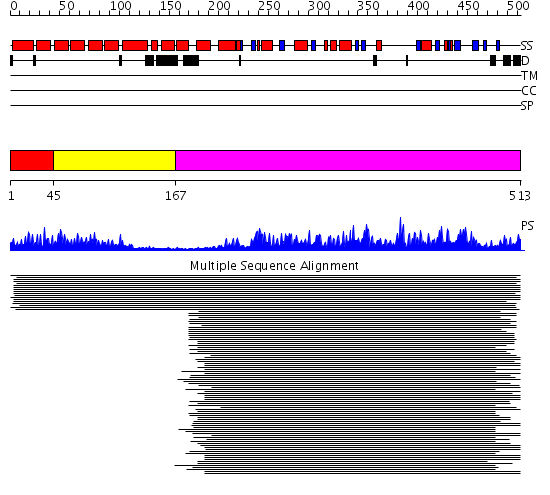
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..44] | 262.440336 | Protein phosphatase 5 | |
| 2 | View Details | [45..166] | 262.440336 | Protein phosphatase 5 | |
| 3 | View Details | [167..513] | 628.0 | Protein phosphatase-1 (PP-1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)