













| General Information: |
|
| Name(s) found: |
BUD3 /
YCL014W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck contractile ring
[TAS cellular bud neck [IDA |
| Biological Process: |
axial cellular bud site selection
[TAS cellular bud site selection [TAS cytokinesis [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKDLSSLYS EKKDKENDET LFNIKLSKSV VETTPLNGHS LFDDDKSLSD WTDNVFTQSV 60
61 FYHGSDDLIW GKFFVCVYKS PNSNKLNAII FDKLGTSCFE SVDISSNSQY YPAIENLSPS 120
121 DQESNVKKCI AVILLQRYPL LSPSDLSQIL SNKSENCDYD PPYAGDLASS CQLITAVPPE 180
181 DLGKRFFTSG LLQNRFVSST LLDVIYENNE STIELNNRLV FHLGEQLEQL FNPVTEYSPE 240
241 QTEYGYKAPE DELPTESDDD LVKAICNELL QLQTNFTFNL VEFLQKFLIA LRVRVLNEEI 300
301 NGLSTTKLNR LFPPTIDEVT RINCIFLDSL KTAIPYGSLE VLKACSITIP YFYKAYTRHE 360
361 AATKNFSKDI KLFIRHFSNV IPEREVYTEM KIESIIKGPQ EKLLKLKLII ERLWKSKKWR 420
421 PKNQEMAKKC YNNIIDVIDS FGKLDSPLHS YSTRVFTPSG KILTELAKCW PVELQYKWLK 480
481 RRVVGVYDVV DLNDENKRNL LVIFSDYVVF INILEAESYY TSDGSNRPLI SDILMNSLIN 540
541 EVPLPSKIPK LKVERHCYID EVLVSILDKS TLRFDRLKGK DSFSMVCKLS SAFISSSSVA 600
601 DLITKARILE KDTAFHLFKA SRSHFTLYST AHELCAYDSE KIKSKFALFL NIPPSKEILE 660
661 VNNLHLAFFA RFCSNDGRDN IVILDVLTKH DDKHIEVTSD NIVFTIINQL AIEIPICFSS 720
721 LNSSMAKDLL CVNENLIKNL EHQLEEVKHP STDEHRAVNS KLSGASDFDA THEKKRSYGT 780
781 ITTFRSYTSD LKDSPSGDNS NVTKETKEIL PVKPTKKSSK KPREIQKKTK TNASKAEHIE 840
841 KKKPNKGKGF FGVLKNVFGS KSKSKPSPVQ RVPKKISQRH PKSPVKKPMT SEKKSSPKRA 900
901 VVSSPKIKKK STSFSTKESQ TAKSSLRAVE FKSDDLIGKP PDVGNGAHPQ ENTRISSVVR 960
961 DTKYVSYNPS QPVTENTSNE KNVEPKADQS TKQDNISNFA DVEVSASSYP EKLDAETDDQ1020
1021 IIGKATNSSS VHGNKELPDL AEVTTANRVS TTSAGDQRID TQSEFLRAAD VENLSDDDEH1080
1081 RQNESRVFND DLFGDFIPKH YRNKQENINS SSNLFPEGKV PQEKGVSNEN TNISLKTNED1140
1141 ASTLTQKLSP QASKVLTENS NELKDTNNEG KDAKDIKLGD DYSDKETAKE ITKPKNFVEG1200
1201 ITERKEIFPT IPRLAPPASK INFQRSPSYI ELFQGMRVVL DKHDAHYNWK RLASQVSLSE1260
1261 GLKVNTEEDA AIINKSQDDA KAERMTQISE VIEYEMQQPI PTYLPKAHLD DSGIEKSDDK1320
1321 FFEIEEELKE ELKGSKTGNE DVGNNNPSNS IPKIEKPPAF KVIRTSPVRI IGRTFEDTRK1380
1381 YENGSPSDIS FTYDTHNNDE PDKRLMELKF PSQDEIPDDR FYTPAEEPTA EFPVEELPNT1440
1441 PRSINVTTSN NKSTDDKLSS GNIDQKPTEL LDDLEFSSFN IAFGNTSMST DNMKISSDLS1500
1501 SNKTVLGNAQ KVQESPSGPL IYVLPQSSTK HEKEGFLRKK QKDEPIWVSP SKIDFADLSR1560
1561 RTKALTPERN TVPLKNNDSR KYKYTGEGSI GNMTNMLLTK DASYAYLKDF VALSDDEDED1620
1621 GKQNCAVGGP EKLKFY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BUD3 HXK1 MRPL20 TDH1 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2006) |
|
ATG26 BUD3 MRPL10 MRPL16 MRPL23 MRPL4 MRPL51 SEC1 |
| View Details | Gavin AC, et al. (2002) |

|
ATG26 BEM2 BUD3 COP1 CYR1 HRR25 IMG1 IMG2 IML1 MHR1 MRH4 MRM1 MRP20 MRP49 MRP7 MRPL1 MRPL10 MRPL13 MRPL16 MRPL17 MRPL19 MRPL20 MRPL23 MRPL24 MRPL25 MRPL27 MRPL28 MRPL3 MRPL35 MRPL36 MRPL39 MRPL4 MRPL44 MRPL51 MRPL6 MRPL7 MRPL8 NUG1 PRP43 RET2 ROM2 RTC6 SEC1 SEC26 SEC27 SIN4 SNF2 SWI3 SYP1 TAF5 UTP20 YDR115W YML6 |
| View Details | Ho Y, et al. (2002) |

|
BRX1 BUD3 CCT2 CCT3 CCT6 CIN8 CLU1 HIR1 HTB1 HTB2 IPI3 KRE33 MNP1 NAN1 NOP56 POL5 RIX1 RPC19 RRP12 RVB1 SSD1 TIF4631 TMA19 UTP10 UTP22 UTP7 |
| View Details | Qiu et al. (2008) |
|
BNR1 BUD3 BUD4 HOF1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
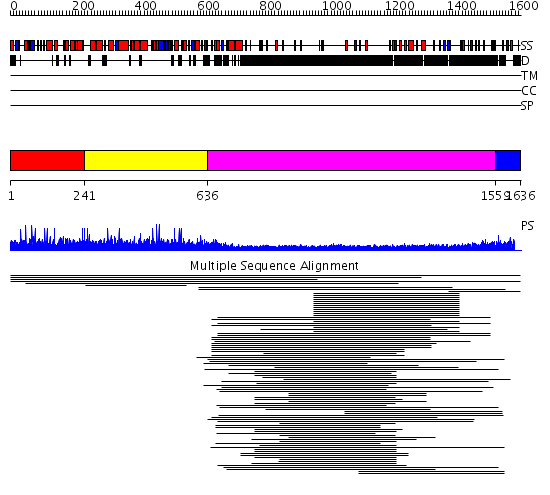
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [241..635] | 1.006863 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [636..1558] | 27.337997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1559..1636] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1038..1216] | 1.422993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1217..1278] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1279..1534] | 1.393995 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1535..1636] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)