













| General Information: |
|
| Name(s) found: |
PSE1 /
YMR308C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1089 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
mRNA export from nucleus
[TAS protein import into nucleus [IMP |
| Molecular Function: |
protein transmembrane transporter activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSALPEEVNR TLLQIVQAFA SPDNQIRSVA EKALSEEWIT ENNIEYLLTF LAEQAAFSQD 60
61 TTVAALSAVL FRKLALKAPP SSKLMIMSKN ITHIRKEVLA QIRSSLLKGF LSERADSIRH 120
121 KLSDAIAECV QDDLPAWPEL LQALIESLKS GNPNFRESSF RILTTVPYLI TAVDINSILP 180
181 IFQSGFTDAS DNVKIAAVTA FVGYFKQLPK SEWSKLGILL PSLLNSLPRF LDDGKDDALA 240
241 SVFESLIELV ELAPKLFKDM FDQIIQFTDM VIKNKDLEPP ARTTALELLT VFSENAPQMC 300
301 KSNQNYGQTL VMVTLIMMTE VSIDDDDAAE WIESDDTDDE EEVTYDHARQ ALDRVALKLG 360
361 GEYLAAPLFQ YLQQMITSTE WRERFAAMMA LSSAAEGCAD VLIGEIPKIL DMVIPLINDP 420
421 HPRVQYGCCN VLGQISTDFS PFIQRTAHDR ILPALISKLT SECTSRVQTH AAAALVNFSE 480
481 FASKDILEPY LDSLLTNLLV LLQSNKLYVQ EQALTTIAFI AEAAKNKFIK YYDTLMPLLL 540
541 NVLKVNNKDN SVLKGKCMEC ATLIGFAVGK EKFHEHSQEL ISILVALQNS DIDEDDALRS 600
601 YLEQSWSRIC RILGDDFVPL LPIVIPPLLI TAKATQDVGL IEEEEAANFQ QYPDWDVVQV 660
661 QGKHIAIHTS VLDDKVSAME LLQSYATLLR GQFAVYVKEV MEEIALPSLD FYLHDGVRAA 720
721 GATLIPILLS CLLAATGTQN EELVLLWHKA SSKLIGGLMS EPMPEITQVY HNSLVNGIKV 780
781 MGDNCLSEDQ LAAFTKGVSA NLTDTYERMQ DRHGDGDEYN ENIDEEEDFT DEDLLDEINK 840
841 SIAAVLKTTN GHYLKNLENI WPMINTFLLD NEPILVIFAL VVIGDLIQYG GEQTASMKNA 900
901 FIPKVTECLI SPDARIRQAA SYIIGVCAQY APSTYADVCI PTLDTLVQIV DFPGSKLEEN 960
961 RSSTENASAA IAKILYAYNS NIPNVDTYTA NWFKTLPTIT DKEAASFNYQ FLSQLIENNS1020
1021 PIVCAQSNIS AVVDSVIQAL NERSLTEREG QTVISSVKKL LGFLPSSDAM AIFNRYPADI1080
1081 MEKVHKWFA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
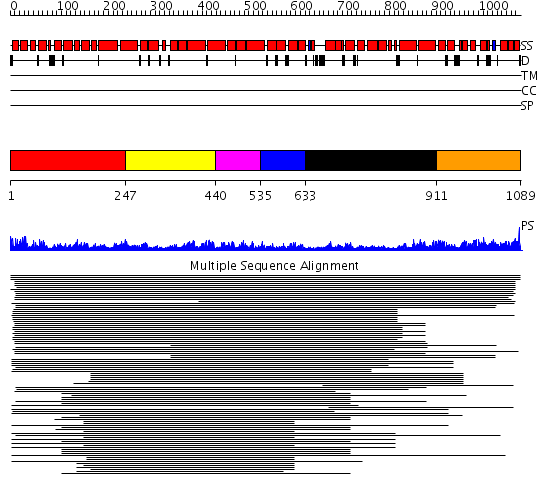
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..246] | 83.39794 | Importin beta | |
| 2 | View Details | [247..439] | 83.39794 | Importin beta | |
| 3 | View Details | [440..534] | 83.39794 | Importin beta | |
| 4 | View Details | [535..632] | 83.39794 | Importin beta | |
| 5 | View Details | [633..910] | 83.39794 | Importin beta | |
| 6 | View Details | [911..1089] | 2.151995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)