













| General Information: |
|
| Name(s) found: |
LYS14 /
YDR034C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
lysine biosynthetic process via aminoadipic acid
[TAS |
| Molecular Function: |
transcription activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFESVNLDEN SPEDRELAKV LSPPGSYLSP ASLDSGSSFT NSGTSTSCFE PKNNLPSLSF 60
61 LNARAGSLGG IFNHKQMTSP SNSNIGGENV ESTTSSNDGS NENAGHPTTS EQDQNADHPT 120
121 ISQADDNGHS SLTPNPAVTS TVTDKKGNTV KRKYSRNGCS ECKRRRMKCD ETKPTCWQCA 180
181 RLNRQCVYVL NPKNKKRRTS NAQRVKEFRK HSTSLDNDHN NARKRQHSSC KAEKKKKVRQ 240
241 NLSEDTTDPK PITDNGKNVP LDEIESLEIP NLDLTTTMNG YDVNLLMQNL NDMVNMKLHD 300
301 SYLLNEELKG LDLPDLDIPE LLPASNVNSS VPISFLVNNV ITFNTKLSSF KLGGIHDKYL 360
361 KIFYYDCLDS IAPFFQNQGN PLRDILLSFA KNEAYLLSSI LATGASIAYR KSNNLEDERN 420
421 YCAYLSHCLS LLGEQFKNES NVLNRIEPII LTVIMLAWDC IYSMNSQWRS HLKGVTDLFK 480
481 KINAGNSSKV LNVAKCWFKV METFASISTV FGGSLIDNND LDAIFDPYDY QYVDSLKFLN 540
541 IMTPLNEFNL LRGHKEDFDL VIKEVFKSLN TIRSTEKNYF SKEEGLFTKK LDYLLLSSQT 600
601 SSEKSKDQIS YFNTQKILVE IDKQLDYEFI DKSGIIPSDN QSHPRISNIH DNAIDMVTLK 660
661 NGEEVAISWY DISHQTQVLS FLLIVLLKLL GMPKESSTIQ QVVKKIMSFF KFLDSDSPPQ 720
721 NSRTCYSNFA VLIAGLNAMD EETRAIVKRY YKINGGKFQK LTEHNLNRLE KVWYGKNQNY 780
781 RLEEQDVLTW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CHS3 LYS14 MSL1 NMD2 SUR7 UPF3 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
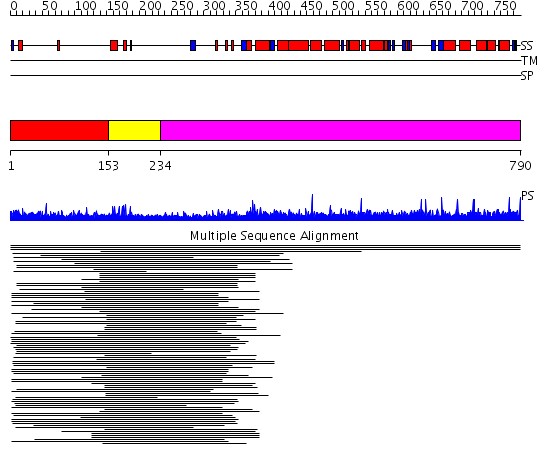
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 21.277997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [153..233] | 5.619789 | Fungal Zn(2)-Cys(6) binuclear cluster domain No confident structure predictions are available. | |
| 3 | View Details | [234..790] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [355..553] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [554..639] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [640..790] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)