













| General Information: |
|
| Name(s) found: |
UPF3 /
YGR072W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 387 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polysome
[IDA nucleus [IDA cytoplasm [TAS |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[TAS telomere maintenance [IMP [NOT]translational frameshifting [IMP mRNA catabolic process [TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNVAGELKN SEGKKKGRGN RYHNKNRGKS KNETVDPKKN ENKVNNATNA THNNSKGRRN 60
61 NKKRNREYYN YKRKARLGKS TENEGFKLVI RLLPPNLTAD EFFAILRDNN NDDGDKQDIQ 120
121 GKLKYSDWCF FEGHYSSKVF KNSTYSRCNF LFDNLSDLEK CANFIKTCKF IDNKDNITIP 180
181 DMKLSPYVKK FTQTSKKDAA LVGTIEEDEI FKTFMNSMKQ LNENDEYSFQ DFSVLKSLEK 240
241 EFSKSIELEN KIAERTERVL TELVGTGDKV KNKNKKKKNK NAKKKFKEEE ASAKIPKKKR 300
301 NRGKKKRENR EKSTISKTKN SNVVIIEEAG KEVLKQRKKK MLLQEKLKIS NSSQPQSSSA 360
361 QTQPSFQPKE NLFVPRVKIL HRDDTKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
NAM7 NMD2 UPF3 |
| View Details | Krogan NJ, et al. (2006) |
|
CHS3 LYS14 MSL1 NMD2 SUR7 UPF3 |
| View Details | Qiu et al. (2008) |

|
NAM7 UPF3 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
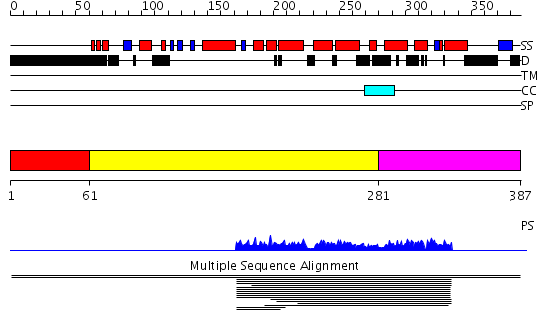
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [61..280] | 121.070581 | Smg-4/UPF3 family No confident structure predictions are available. | |
| 3 | View Details | [281..387] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [303..387] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)