













| General Information: |
|
| Name(s) found: |
RFC1 /
YOR217W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 861 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA replication factor C complex
[IDA |
| Biological Process: |
DNA repair
[IMP leading strand elongation [IDA mismatch repair [TAS |
| Molecular Function: |
DNA clamp loader activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNISDFFGK NKKSVRSSTS RPTRQVGSSK PEVIDLDTES DQESTNKTPK KMPVSNVIDV 60
61 SETPEGEKKL PLPAKRKASS PTVKPASSKK TKPSSKSSDS ASNITAQDVL DKIPSLDLSN 120
121 VHVKENAKFD FKSANSNADP DEIVSEIGSF PEGKPNCLLG LTIVFTGVLP TLERGASEAL 180
181 AKRYGARVTK SISSKTSVVV LGDEAGPKKL EKIKQLKIKA IDEEGFKQLI AGMPAEGGDG 240
241 EAAEKARRKL EEQHNIATKE AELLVKKEEE RSKKLAATRV SGGHLERDNV VREEDKLWTV 300
301 KYAPTNLQQV CGNKGSVMKL KNWLANWENS KKNSFKHAGK DGSGVFRAAM LYGPPGIGKT 360
361 TAAHLVAQEL GYDILEQNAS DVRSKTLLNA GVKNALDNMS VVGYFKHNEE AQNLNGKHFV 420
421 IIMDEVDGMS GGDRGGVGQL AQFCRKTSTP LILICNERNL PKMRPFDRVC LDIQFRRPDA 480
481 NSIKSRLMTI AIREKFKLDP NVIDRLIQTT RGDIRQVINL LSTISTTTKT INHENINEIS 540
541 KAWEKNIALK PFDIAHKMLD GQIYSDIGSR NFTLNDKIAL YFDDFDFTPL MIQENYLSTR 600
601 PSVLKPGQSH LEAVAEAANC ISLGDIVEKK IRSSEQLWSL LPLHAVLSSV YPASKVAGHM 660
661 AGRINFTAWL GQNSKSAKYY RLLQEIHYHT RLGTSTDKIG LRLDYLPTFR KRLLDPFLKQ 720
721 GADAISSVIE VMDDYYLTKE DWDSIMEFFV GPDVTTAIIK KIPATVKSGF TRKYNSMTHP 780
781 VAIYRTGSTI GGGGVGTSTS TPDFEDVVDA DDNPVPADDE ETQDSSTDLK KDKLIKQKAK 840
841 PTKRKTATSK PGGSKKRKTK A |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
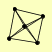
|
RFC1 RFC2 RFC3 RFC4 RFC5 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
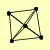
|
RFC1 RFC2 RFC3 RFC4 RFC5 |
| View Details | Krogan NJ, et al. (2006) |
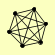
|
ELG1 RFC1 RFC2 RFC3 RFC4 RFC5 |
| View Details | Gavin AC, et al. (2006) |
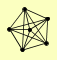
|
ELG1 RFC1 RFC2 RFC3 RFC4 RFC5 |
| View Details | Gavin AC, et al. (2002) |

|
CTF18 CTF8 DPB2 ELG1 MIS1 OLA1 POL1 POL12 POL2 PRI1 PRI2 RFC1 RFC2 RFC3 RFC4 RFC5 TOP2 USO1 VPS13 YMR181C |
| View Details | Ho Y, et al. (2002) |
|
ACO1 ADE5,7 ADH2 AHA1 ELG1 GUS1 HSP104 RFC1 RFC2 RFC4 RNQ1 RPT3 SAN1 YHR020W |
| View Details | Qiu et al. (2008) |
|
CDC45 DPB11 DPB2 DPB4 MCM2 MCM3 MCM4 MCM6 MCM7 POL3 POL31 POL32 RFC1 RFC2 RFC3 RFC4 RFC5 RRM3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
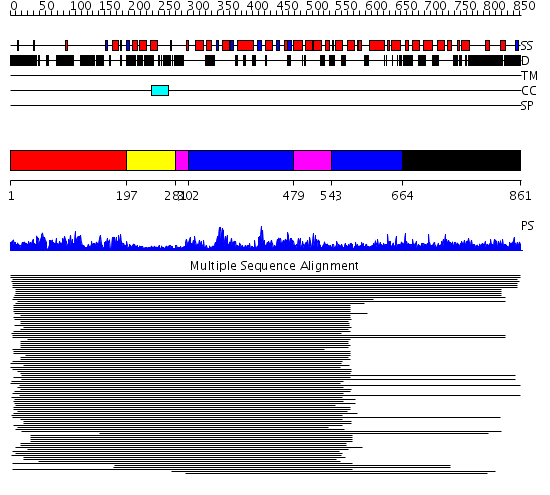
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 70.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [197..280] | 70.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [281..301] [479..542] |
207.06601 | Replication factor C | |
| 4 | View Details | [302..478] [543..663] |
207.06601 | Replication factor C | |
| 5 | View Details | [664..861] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)