













| General Information: |
|
| Name(s) found: |
TSL1 /
YML100W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1098 amino acids |
Gene Ontology: |
|
| Cellular Component: |
alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)
[IGI |
| Biological Process: |
trehalose biosynthetic process
[IGI response to stress [IMP |
| Molecular Function: |
alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity
[IMP trehalose-phosphatase activity [IGI enzyme regulator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALIVASLFL PYQPQFELDT SLPENSQVDS SLVNIQAMAN DQQQQRALSN NISQESLVAP 60
61 APEQGVPPAI SRSATRSPSA FNRASSTTNT ATLDDLVSSD IFMENLTANA TTSHTPTSKT 120
121 MLKPRKNGSV ERFFSPSSNI PTDRIASPIQ HEHDSGSRIA SPIQQQQQDP TTNLLKNVNK 180
181 SLLVHSLLNN TSQTSLEGPN NHIVTPKSRA GNRPTSAATS LVNRTKQGSA SSGSSGSSAP 240
241 PSIKRITPHL TASAAKQRPL LAKQPSNLKY SELADISSSE TSSQHNESDP DDLTTAPDEE 300
301 YVSDLEMDDA KQDYKVPKFG GYSNKSKLKK YALLRSSQEL FSRLPWSIVP SIKGNGAMKN 360
361 AINTAVLENI IPHRHVKWVG TVGIPTDEIP ENILANISDS LKDKYDSYPV LTDDDTFKAA 420
421 YKNYCKQILW PTLHYQIPDN PNSKAFEDHS WKFYRNLNQR FADAIVKIYK KGDTIWIHDY 480
481 HLMLVPQMVR DVLPFAKIGF TLHVSFPSSE VFRCLAQREK ILEGLTGADF VGFQTREYAR 540
541 HFLQTSNRLL MADVVHDEEL KYNGRVVSVR FTPVGIDAFD LQSQLKDGSV MQWRQLIRER 600
601 WQGKKLIVCR DQFDRIRGIH KKLLAYEKFL VENPEYVEKS TLIQICIGSS KDVELERQIM 660
661 IVVDRINSLS TNISISQPVV FLHQDLDFSQ YLALSSEADL FVVSSLREGM NLTCHEFIVC 720
721 SEDKNAPLLL SEFTGSASLL NDGAIIINPW DTKNFSQAIL KGLEMPFDKR RPQWKKLMKD 780
781 IINNDSTNWI KTSLQDIHIS WQFNQEGSKI FKLNTKTLME DYQSSKKRMF VFNIAEPPSS 840
841 RMISILNDMT SKGNIVYIMN SFPKPILENL YSRVQNIGLI AENGAYVSLN GVWYNIVDQV 900
901 DWRNDVAKIL EDKVERLPGS YYKINESMIK FHTENAEDQD RVASVIGDAI THINTVFDHR 960
961 GIHAYVYKNV VSVQQVGLSL SAAQFLFRFY NSASDPLDTS SGQITNIQTP SQQNPSDQEQ1020
1021 QPPASPTVSM NHIDFACVSG SSSPVLEPLF KLVNDEASEG QVKAGHAIVY GDATSTYAKE1080
1081 HVNGLNELFT IISRIIED |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
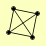
|
TPS1 TPS2 TPS3 TSL1 |
| View Details | Krogan NJ, et al. (2006) |
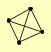
|
TPS1 TPS2 TPS3 TSL1 |
| View Details | Gavin AC, et al. (2002) |
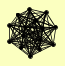
|
CTR3 DAL3 GLN1 GTO3 IML1 MCM2 MKS1 REG1 TIF1, TIF2 TPS1 TPS2 TPS3 TSC10 TSL1 YIL177C |
| View Details | Ho Y, et al. (2002) |
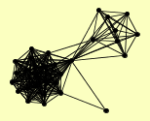
|
CDC25 CKI1 DSK2 GCR2 GIN4 HMF1 HOM3 IRA1 IRA2 MYO2 MYO4 NAP1 NPL4 PMD1 PRS2 PRS3 PRS5 PTC1 RAD23 RIM11 SHP1 TPS1 TSL1 UBI4 UFD2 YDR049W YDR170W-A YER138C YER160C YJR027W YJR028W |
| View Details | Qiu et al. (2008) |
|
GIP2 REG1 TPS1 TPS2 TSL1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
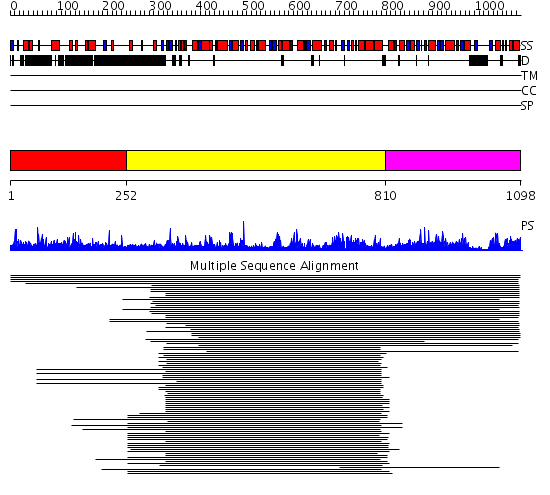
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | 2.067997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [252..809] | 248.022276 | Glycosyltransferase family 20 No confident structure predictions are available. | |
| 3 | View Details | [810..1098] | 73.136677 | Trehalose-phosphatase No confident structure predictions are available. | |
| 4 | View Details | [814..1098] | 10.09691 | Crystal structure of trehalose-6-phosphate phosphatase related protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)