













| General Information: |
|
| Name(s) found: |
GCN2 /
YDR283C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1659 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic ribosome
[IDA]
|
| Biological Process: |
cellular amino acid biosynthetic process
[IMP regulation of translational initiation [IDA protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein kinase activity
[IDA eukaryotic translation initiation factor 2alpha kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSHLTLDQ YYEIQCNELE AIRSIYMDDF TDLTKRKSSW DKQPQIIFEI TLRSVDKEPV 60
61 ESSITLHFAM TPMYPYTAPE IEFKNVQNVM DSQLQMLKSE FKKIHNTSRG QEIIFEITSF 120
121 TQEKLDEFQN VVNTQSLEDD RLQRIKETKE QLEKEEREKQ QETIKKRSDE QRRIDEIVQR 180
181 ELEKRQDDDD DLLFNRTTQL DLQPPSEWVA SGEAIVFSKT IKAKLPNNSM FKFKAVVNPK 240
241 PIKLTSDIFS FSKQFLVKPY IPPESPLADF LMSSEMMENF YYLLSEIELD NSYFNTSNGK 300
301 KEIANLEKEL ETVLKAKHDN VNRLFGYTVE RMGRNNATFV WKIRLLTEYC NYYPLGDLIQ 360
361 SVGFVNLATA RIWMIRLLEG LEAIHKLGIV HKCINLETVI LVKDADFGST IPKLVHSTYG 420
421 YTVLNMLSRY PNKNGSSVEL SPSTWIAPEL LKFNNAKPQR LTDIWQLGVL FIQIISGSDI 480
481 VMNFETPQEF LDSTSMDETL YDLLSKMLNN DPKKRLGTLE LLPMKFLRTN IDSTINRFNL 540
541 VSESVNSNSL ELTPGDTITV RGNGGRTLSQ SSIRRRSFNV GSRFSSINPA TRSRYASDFE 600
601 EIAVLGQGAF GQVVKARNAL DSRYYAIKKI RHTEEKLSTI LSEVMLLASL NHQYVVRYYA 660
661 AWLEEDSMDE NVFESTDEES DLSESSSDFE ENDLLDQSSI FKNRTNHDLD NSNWDFISGS 720
721 GYPDIVFENS SRDDENEDLD HDTSSTSSSE SQDDTDKESK SIQNVPRRRN FVKPMTAVKK 780
781 KSTLFIQMEY CENRTLYDLI HSENLNQQRD EYWRLFRQIL EALSYIHSQG IIHRDLKPMN 840
841 IFIDESRNVK IGDFGLAKNV HRSLDILKLD SQNLPGSSDN LTSAIGTAMY VATEVLDGTG 900
901 HYNEKIDMYS LGIIFFEMIY PFSTGMERVN ILKKLRSVSI EFPPDFDDNK MKVEKKIIRL 960
961 LIDHDPNKRP GARTLLNSGW LPVKHQDEVI KEALKSLSNP SSPWQQQVRE SLFNQSYSLT1020
1021 NDILFDNSVP TSTPFANILR SQMTEEVVKI FRKHGGIENN APPRIFPKAP IYGTQNVYEV1080
1081 LDKGGTVLQL QYDLTYPMAR YLSKNPSLIS KQYRMQHVYR PPDHSRSSLE PRKFGEIDFD1140
1141 IISKSSSESG FYDAESLKII DEILTVFPVF EKTNTFFILN HADILESVFN FTNIDKAQRP1200
1201 LVSRMLSQVG FARSFKEVKN ELKAQLNISS TALNDLELFD FRLDFEAAKK RLYKLMIDSP1260
1261 HLKKIEDSLS HISKVLSYLK PLEVARNVVI SPLSNYNSAF YKGGIMFHAV YDDGSSRNMI1320
1321 AAGGRYDTLI SFFARPSGKK SSNTRKAVGF NLAWETIFGI AQNYFKLASG NRIKKRNRFL1380
1381 KDTAVDWKPS RCDVLISSFS NSLLDTIGVT ILNTLWKQNI KADMLRDCSS VDDVVTGAQQ1440
1441 DGIDWILLIK QQAYPLTNHK RKYKPLKIKK LSTNVDIDLD LDEFLTLYQQ ETGNKSLIND1500
1501 SLTLGDKADE FKRWDENSSA GSSQEGDIDD VVAGSTNNQK VIYVPNMATR SKKANKREKW1560
1561 VYEDAARNSS NMILHNLSNA PIITVDALRD ETLEIISITS LAQKEEWLRK VFGSGNNSTP1620
1621 RSFATSIYNN LSKEAHKGNR WAILYCHKTG KSSVIDLQR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
AAR2 GCN2 LTV1 RPM2 SNU66 |
| View Details | Gavin AC, et al. (2002) |

|
AAR2 BRR1 BUD31 CDC33 CEF1 CLF1 CSN12 CUS1 CWC2 CWC22 CWC23 DHH1 DIB1 ECM2 GCN2 HSH155 HTA1 IMG1 LEA1 LIN1 LSM1 LSM2 LSM4 LSM5 LSM6 LSM7 LUC7 MRP7 MRPL28 MRPL3 MRPL35 MRPL38 MRPL4 MRPL8 MRPS5 MSL1 MUD1 PAT1 PRP11 PRP18 PRP19 PRP21 PRP24 PRP28 PRP3 PRP31 PRP38 PRP39 PRP4 PRP40 PRP42 PRP43 PRP46 PRP6 PRP8 PRP9 RSE1 RVB2 SLU7 SMB1 SMD1 SMD2 SMD3 SME1 SMX2 SMX3 SNP1 SNT309 SNU114 SNU23 SNU56 SNU66 SNU71 SPP381 SPP382 SRB2 STO1 SYF1 TOS4 UTP21 YML6 |
| View Details | Ho Y, et al. (2002) |

|
GCN2 RRG9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
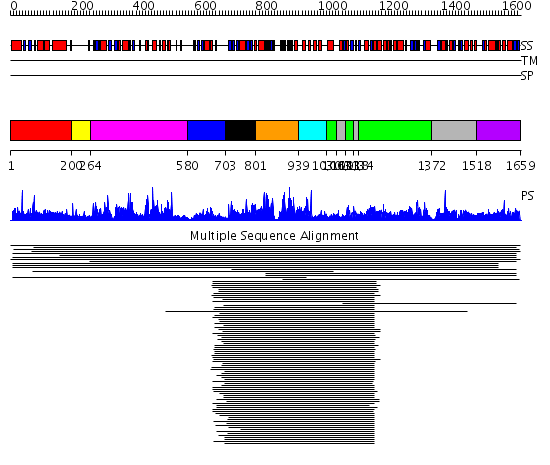
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 1.054963 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [200..263] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [264..579] | 160.626727 | MAP kinase Erk2 | |
| 4 | View Details | [580..702] | 57.201249 | No description for 1lwpA_ was found. | |
| 5 | View Details | [703..800] | 57.201249 | No description for 1lwpA_ was found. | |
| 6 | View Details | [801..938] | 57.201249 | No description for 1lwpA_ was found. | |
| 7 | View Details | [939..1029] | 271.0 | Calmodulin-dependent protein kinase | |
| 8 | View Details | [1030..1062] [1093..1117] [1134..1371] |
16.0 | Histidyl-tRNA synthetase (HisRS), C-terminal domain; Histidyl-tRNA synthetase (HisRS) | |
| 9 | View Details | [1063..1092] [1118..1133] [1372..1517] |
16.0 | Histidyl-tRNA synthetase (HisRS), C-terminal domain; Histidyl-tRNA synthetase (HisRS) | |
| 10 | View Details | [1518..1659] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)