













| General Information: |
|
| Name(s) found: |
CNS1 /
YBR155W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 385 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IPI |
| Biological Process: |
protein folding
[IGI |
| Molecular Function: |
Hsp90 protein binding
[IDA Hsp70 protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVNANGGY TKPQKYVPGP GDPELPPQLS EFKDKTSDEI LKEMNRMPFF MTKLDETDGA 60
61 GGENVELEAL KALAYEGEPH EIAENFKKQG NELYKAKRFK DARELYSKGL AVECEDKSIN 120
121 ESLYANRAAC ELELKNYRRC IEDCSKALTI NPKNVKCYYR TSKAFFQLNK LEEAKSAATF 180
181 ANQRIDPENK SILNMLSVID RKEQELKAKE EKQQREAQER ENKKIMLESA MTLRNITNIK 240
241 THSPVELLNE GKIRLEDPMD FESQLIYPAL IMYPTQDEFD FVGEVSELTT VQELVDLVLE 300
301 GPQERFKKEG KENFTPKKVL VFMETKAGGL IKAGKKLTFH DILKKESPDV PLFDNALKIY 360
361 IVPKVESEGW ISKWDKQKAL ERRSV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2002) |
|
CNS1 HGH1 |
| View Details | Ho Y, et al. (2002) |
|
CNS1 ECM10 ILV5 YHB1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
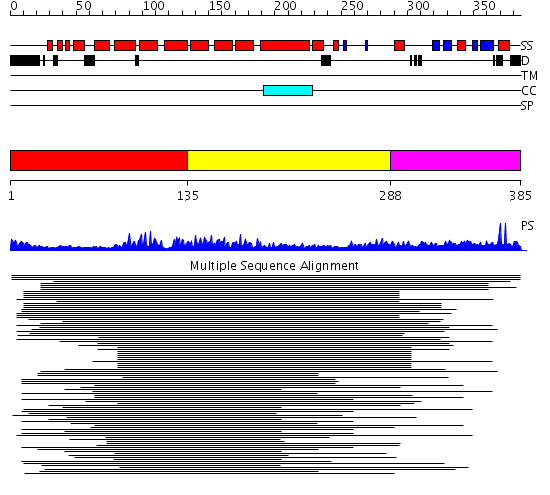
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 103.211549 | Cyclophilin 40; Cyclophilin 40 isomerase domain | |
| 2 | View Details | [135..287] | 103.211549 | Cyclophilin 40; Cyclophilin 40 isomerase domain | |
| 3 | View Details | [288..385] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)