













| General Information: |
|
| Name(s) found: |
RAD61 /
YDR014W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS |
| Biological Process: |
response to radiation
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRAYGKRGPV LRTPFRSNKG LPSSSDVEFS DDDVNSVIPD VSSTISSSIA DHPIEGLLDE 60
61 PRKAQDSSSS FDGANEKPSS QLDSKRNDQN VKIITSSDTS MAFMKDEKLS AFNFLDGSKA 120
121 SKRKRRRTYQ KHDANITSSI EPDVQDEDSI TMHNEFESIR KIYNDINEFI LKLPRADDDI 180
181 LNKMLENEMK MDDSIENNSI RTSKDKKYGK FRTILINKNK ENEIMGEEVD QKANTLSLNN 240
241 ADNSNAEKEG LTSTNHYNEL KNMGDTIKYQ DDIEFLLSNS KSNDNTTVPI NEYFKKLLNL 300
301 SLMIINDEEF FQYAKRYFKK EIIKLSFAQF RSDFPELILL QGYLLHKVSE SQSDFPPSFD 360
361 NFSIELSKDD GKIRTKKNKH IKKLSHLNFE DFLRKTQFKT GLYYSLSLWE MHGNLSLDII 420
421 KRISILASNK DLFSRHVKTF IPLLEKLITA SEFCHMYIEQ PEMFDSLISN LNNQFKDMLD 480
481 DDSLIKILIL LTNMEVHNYT LWKEADMIFQ SSMNTILESI HPLTDAKVDN ILLHLGLCLN 540
541 ICSRENSRLK LDGKLWYDMK TIFVKMIRDG SDTENRLVQG LFYLNFSFLI KQRKENSNLD 600
601 PGELNLLLVE LEAFKSETSQ FNEGISNKIE IALNYLKSIY TSERITI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
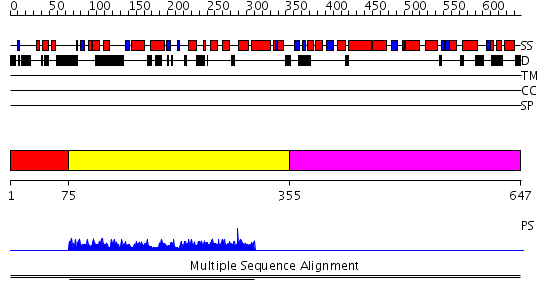
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..354] | 1.00292 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [355..647] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)