













| General Information: |
|
| Name(s) found: |
YNK1 /
YKL067W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 153 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial intermembrane space
[IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
RNA metabolic process
[IDA purine nucleotide biosynthetic process [TAS nucleoside triphosphate biosynthetic process [IDA nucleoside diphosphate phosphorylation [IDA DNA metabolic process [IDA nucleotide metabolic process [TAS |
| Molecular Function: |
nucleoside diphosphate kinase activity
[IDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ADE3 DPS1 FUR1 RPL30 SAR1 TDH1 YDR341C YNK1 |
| View Details | Gavin AC, et al. (2002) |
|
HHF1, HHF2 RPA135 YNK1 |
| View Details | Ho Y, et al. (2002) |

|
AAT2 ACH1 ACO1 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CCT5 CDC15 CDC33 CLU1 CMD1 CMK2 CMP2 COF1 COR1 CPR1 CYR1 CYS4 DUG2 DUN1 EDE1 EGD1 ERG13 ERG6 FPR1 FRS2 GCD11 GND1 GND2 GRX1 GUS1 HCH1 HEF3 HHF1, HHF2 HYP2 ILS1 IPP1 LRO1 LSP1 MCX1 MET6 MYO2 MYO4 MYO5 NAP1 NMD3 NTF2 OLA1 OYE2 PFK1 PGM2 PIL1 PRM2 PRP19 PST2 QCR2 RNR2 RSN1 SAH1 SAR1 SCP160 SEC53 SEN15 SES1 SHE3 SNU13 SOD1 TCP1 TEF4 TEM1 THS1 TIF1, TIF2 TMA19 TSA2 UBA1 VAS1 VMA4 VMA5 VPS13 WTM1 YHR033W YJU2 YNK1 |
| View Details | Qiu et al. (2008) |
|
ADK1 ADK2 STF2 YNK1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
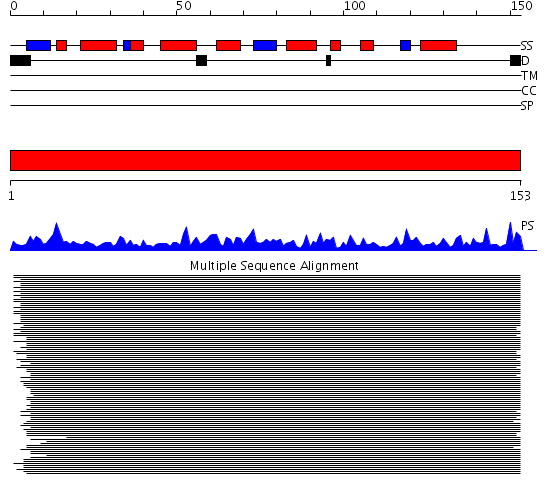
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 596.502279 | Nucleoside diphosphate kinases |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)