













| General Information: |
|
| Name(s) found: |
RFA3 /
YJL173C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 122 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome, telomeric region
[IMP DNA replication factor A complex [TAS |
| Biological Process: |
DNA strand elongation involved in DNA replication
[TAS double-strand break repair via homologous recombination [IGI DNA recombination [TAS nucleotide-excision repair [TAS DNA unwinding involved in replication [TAS postreplication repair [TAS DNA replication, synthesis of RNA primer [TAS |
| Molecular Function: |
DNA binding
[TAS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
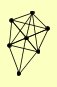
|
DNA2 RAD52 RFA1 RFA2 RFA3 RIM1 RTT105 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
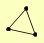
|
RFA1 RFA2 RFA3 |
| View Details | Krogan NJ, et al. (2006) |
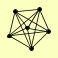
|
DNA2 HSP12 PRO1 RFA1 RFA2 RFA3 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |
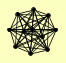
|
AAC3 ARO1 CYS4 HEF3 HOR2 HXT7 MTC5 PGM2 RFA3 RHR2 YJR141W |
| View Details | Qiu et al. (2008) |
|
MCM6 PRI2 RFA1 RFA2 RFA3 RFC4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
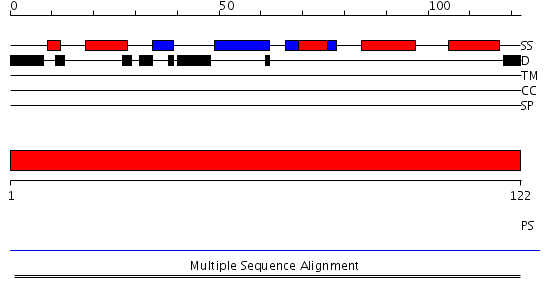
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)