













| General Information: |
|
| Name(s) found: |
PXR1 /
YGR280C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 271 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [IDA |
| Biological Process: |
ribosome biogenesis
[RCA snoRNA 3'-end processing [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLAATRTKQ RFGLDPRNTA WSNDTSRFGH QFLEKFGWKP GMGLGLSPMN SNTSHIKVSI 60
61 KDDNVGLGAK LKRKDKKDEF DNGECAGLDV FQRILGRLNG KESKISEELD TQRKQKIIDG 120
121 KWGIHFVKGE VLASTWDPKT HKLRNYSNAK KRKREGDDSE DEDDDDKEDK DSDKKKHKKH 180
181 KKHKKDKKKD KKDKKEHKKH KKEEKRLKKE KRAEKTKETK KTSKLKSSES ASNIPDAVNT 240
241 RLSVRSKWIK QKRAALMDSK ALNEIFMITN D |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
GDE1 PXR1 YME2 |
| View Details | Qiu et al. (2008) |

|
BRX1 CBF5 DBP3 DBP7 DBP8 DHR2 DRS1 ENP1 ERB1 ESF1 ESF2 FCF2 FPR4 GAR1 HCA4 KRE33 KRI1 KRR1 MAK11 MAK5 MRT4 NOC2 NOC3 NOP12 NOP15 NOP16 NOP2 NOP6 NOP7 NOP8 NOP9 NSA1 NSR1 NUG1 PUF6 PXR1 RPF2 RPL27A RRB1 RRP1 RRP9 SLX9 SOF1 SPB4 SSF1 UTP10 UTP13 UTP14 UTP15 UTP4 UTP8 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
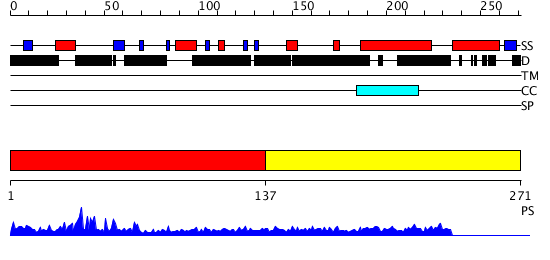
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 13.346787 | G-patch domain No confident structure predictions are available. | |
| 2 | View Details | [137..271] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)