













| General Information: |
|
| Name(s) found: |
YCK3 /
YER123W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 524 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA fungal-type vacuole membrane [IDA plasma membrane [IDA |
| Biological Process: |
protein amino acid phosphorylation
[ISS |
| Molecular Function: |
casein kinase I activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQRSSQHIV GIHYAVGPKI GEGSFGVIFE GENILHSCQA QTGSKRDSSI IMANEPVAIK 60
61 FEPRHSDAPQ LRDEFRAYRI LNGCVGIPHA YYFGQEGMHN ILIIDLLGPS LEDLFEWCGR 120
121 KFSVKTTCMV AKQMIDRVRA IHDHDLIYRD IKPDNFLISQ YQRISPEGKV IKSCASSSNN 180
181 DPNLIYMVDF GMAKQYRDPR TKQHIPYRER KSLSGTARYM SINTHFGREQ SRRDDLESLG 240
241 HVFFYFLRGS LPWQGLKAPN NKLKYEKIGM TKQKLNPDDL LLNNAIPYQF ATYLKYARSL 300
301 KFDEDPDYDY LISLMDDALR LNDLKDDGHY DWMDLNGGKG WNIKINRRAN LHGYGNPNPR 360
361 VNGNTARNNV NTNSKTRNTT PVATPKQQAQ NSYNKDNSKS RISSNPQSFT KQQHVLKKIE 420
421 PNSKYIPETH SNLQRPIKSQ SQTYDSISHT QNSPFVPYSS SKANPKRSNN EHNLPNHYTN 480
481 LANKNINYQS QRNYEQENDA YSDDENDTFC SKIYKYCCCC FCCC |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
HRR25 YCK1 YCK2 YCK3 |
| View Details | Ho Y, et al. (2002) |
|
BCY1 CAR2 CYS4 GND1 GTO1 LAA1 LYS1 SAH1 SNO2 SSN8 THI22 THO2 TIF1, TIF2 TPK2 TPK3 VPS21 YBR028C YCK1 YCK2 YCK3 YGR111W YPT53 |
| View Details | Qiu et al. (2008) |

|
YCK2 YCK3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
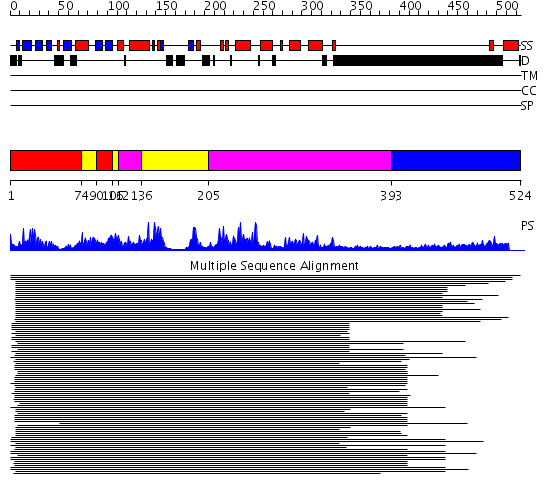
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] [90..105] |
1185.045757 | Casein kinase-1, CK1 | |
| 2 | View Details | [74..89] [106..111] [136..204] |
1185.045757 | Casein kinase-1, CK1 | |
| 3 | View Details | [112..135] [205..392] |
1185.045757 | Casein kinase-1, CK1 | |
| 4 | View Details | [393..524] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)